Identification of a mutant PfCRT-mediated chloroquine tolerance phenotype in Plasmodium falciparum
- PMID: 20485514
- PMCID: PMC2869323
- DOI: 10.1371/journal.ppat.1000887
Identification of a mutant PfCRT-mediated chloroquine tolerance phenotype in Plasmodium falciparum
Abstract
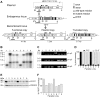
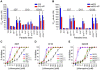
Mutant forms of the Plasmodium falciparum transporter PfCRT constitute the key determinant of parasite resistance to chloroquine (CQ), the former first-line antimalarial, and are ubiquitous to infections that fail CQ treatment. However, treatment can often be successful in individuals harboring mutant pfcrt alleles, raising questions about the role of host immunity or pharmacokinetics vs. the parasite genetic background in contributing to treatment outcomes. To examine whether the parasite genetic background dictates the degree of mutant pfcrt-mediated CQ resistance, we replaced the wild type pfcrt allele in three CQ-sensitive strains with mutant pfcrt of the 7G8 allelic type prevalent in South America, the Oceanic region and India. Recombinant clones exhibited strain-dependent CQ responses that ranged from high-level resistance to an incremental shift that did not meet CQ resistance criteria. Nonetheless, even in the most susceptible clones, 7G8 mutant pfcrt enabled parasites to tolerate CQ pressure and recrudesce in vitro after treatment with high concentrations of CQ. 7G8 mutant pfcrt was found to significantly impact parasite responses to other antimalarials used in artemisinin-based combination therapies, in a strain-dependent manner. We also report clinical isolates from French Guiana that harbor mutant pfcrt, identical or related to the 7G8 haplotype, and manifest a CQ tolerance phenotype. One isolate, H209, harbored a novel PfCRT C350R mutation and demonstrated reduced quinine and artemisinin susceptibility. Our data: 1) suggest that high-level CQR is a complex biological process dependent on the presence of mutant pfcrt; 2) implicate a role for variant pfcrt alleles in modulating parasite susceptibility to other clinically important antimalarials; and 3) uncover the existence of a phenotype of CQ tolerance in some strains harboring mutant pfcrt.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Adaptive evolution of malaria parasites in French Guiana: Reversal of chloroquine resistance by acquisition of a mutation in pfcrt.Proc Natl Acad Sci U S A. 2015 Sep 15;112(37):11672-7. doi: 10.1073/pnas.1507142112. Epub 2015 Aug 10. Proc Natl Acad Sci U S A. 2015. PMID: 26261345 Free PMC article.
-
Combinatorial Genetic Modeling of pfcrt-Mediated Drug Resistance Evolution in Plasmodium falciparum.Mol Biol Evol. 2016 Jun;33(6):1554-70. doi: 10.1093/molbev/msw037. Epub 2016 Feb 22. Mol Biol Evol. 2016. PMID: 26908582 Free PMC article.
-
A Variant PfCRT Isoform Can Contribute to Plasmodium falciparum Resistance to the First-Line Partner Drug Piperaquine.mBio. 2017 May 9;8(3):e00303-17. doi: 10.1128/mBio.00303-17. mBio. 2017. PMID: 28487425 Free PMC article.
-
PfCRT and its role in antimalarial drug resistance.Trends Parasitol. 2012 Nov;28(11):504-14. doi: 10.1016/j.pt.2012.08.002. Epub 2012 Sep 25. Trends Parasitol. 2012. PMID: 23020971 Free PMC article. Review.
-
pfcrt is more than the Plasmodium falciparum chloroquine resistance gene: a functional and evolutionary perspective.Acta Trop. 2005 Jun;94(3):170-80. doi: 10.1016/j.actatropica.2005.04.004. Acta Trop. 2005. PMID: 15866507 Review.
Cited by
-
Degrees of chloroquine resistance in Plasmodium - is the redox system involved?Int J Parasitol Drugs Drug Resist. 2012 Dec 1;2:47-57. doi: 10.1016/j.ijpddr.2011.11.001. Int J Parasitol Drugs Drug Resist. 2012. PMID: 22773965 Free PMC article.
-
Differential drug efflux or accumulation does not explain variation in the chloroquine response of Plasmodium falciparum strains expressing the same isoform of mutant PfCRT.Antimicrob Agents Chemother. 2011 May;55(5):2310-8. doi: 10.1128/AAC.01167-10. Epub 2011 Feb 22. Antimicrob Agents Chemother. 2011. PMID: 21343459 Free PMC article.
-
Discordant temporal evolution of Pfcrt and Pfmdr1 genotypes and Plasmodium falciparum in vitro drug susceptibility to 4-aminoquinolines after drug policy change in French Guiana.Antimicrob Agents Chemother. 2012 Mar;56(3):1382-9. doi: 10.1128/AAC.05280-11. Epub 2012 Jan 9. Antimicrob Agents Chemother. 2012. PMID: 22232280 Free PMC article.
-
In Vitro Antiplasmodium and Chloroquine Resistance Reversal Effects of Andrographolide.Evid Based Complement Alternat Med. 2019 Dec 13;2019:7967980. doi: 10.1155/2019/7967980. eCollection 2019. Evid Based Complement Alternat Med. 2019. PMID: 31915453 Free PMC article.
-
Quantitative assessment of Plasmodium falciparum sexual development reveals potent transmission-blocking activity by methylene blue.Proc Natl Acad Sci U S A. 2011 Nov 22;108(47):E1214-23. doi: 10.1073/pnas.1112037108. Epub 2011 Oct 31. Proc Natl Acad Sci U S A. 2011. PMID: 22042867 Free PMC article.
References
-
- Wellems TE, Plowe CV. Chloroquine-resistant malaria. J Infect Dis. 2001;184:770–776. - PubMed
-
- Wongsrichanalai C, Pickard AL, Wernsdorfer WH, Meshnick SR. Epidemiology of drug-resistant malaria. Lancet Infect Dis. 2002;2:209–218. - PubMed
-
- Rieckmann KH. The chequered history of malaria control: are new and better tools the ultimate answer? Ann Trop Med Parasitol. 2006;100:647–662. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

