A super-resolution map of the vertebrate kinetochore
- PMID: 20483991
- PMCID: PMC2890832
- DOI: 10.1073/pnas.1002325107
A super-resolution map of the vertebrate kinetochore
Abstract
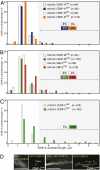
A longstanding question in centromere biology has been the organization of CENP-A-containing chromatin and its implications for kinetochore assembly. Here, we have combined genetic manipulations with deconvolution and super-resolution fluorescence microscopy for a detailed structural analysis of chicken kinetochores. Using fluorescence microscopy with subdiffraction spatial resolution and single molecule sensitivity to map protein localization in kinetochore chromatin unfolded by exposure to a low salt buffer, we observed robust amounts of H3K9me3, but only low levels of H3K4me2, between CENP-A subdomains in unfolded interphase prekinetochores. Constitutive centromere-associated network proteins CENP-C and CENP-H localize within CENP-A-rich subdomains (presumably on H3-containing nucleosomes) whereas CENP-T localizes in interspersed H3-rich blocks. Although interphase prekinetochores are relatively more resistant to unfolding than sur-rounding pericentromeric heterochromatin, mitotic kinetochores are significantly more stable, reflecting mitotic kinetochore maturation. Loss of CENP-H, CENP-N, or CENP-W had little or no effect on the unfolding of mitotic kinetochores. However, loss of CENP-C caused mitotic kinetochores to unfold to the same extent as their interphase counterparts. Based on our results we propose a new model for inner centromeric chromatin architecture in which chromatin is folded as a layered boustrophedon, with planar sinusoids containing interspersed CENP-A-rich and H3-rich subdomains oriented toward the outer kinetochore. In mitosis, a CENP-C-dependent mechanism crosslinks CENP-A blocks of different layers together, conferring extra stability to the kinetochore.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



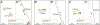

Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Evaluating Different Types of Cancer Survivorship Care [Internet].Washington (DC): Patient-Centered Outcomes Research Institute (PCORI); 2019 Jul. Washington (DC): Patient-Centered Outcomes Research Institute (PCORI); 2019 Jul. PMID: 39602557 Free Books & Documents. Review.
-
The effectiveness of abstinence-based and harm reduction-based interventions in reducing problematic substance use in adults who are experiencing homelessness in high income countries: A systematic review and meta-analysis: A systematic review.Campbell Syst Rev. 2024 Apr 21;20(2):e1396. doi: 10.1002/cl2.1396. eCollection 2024 Jun. Campbell Syst Rev. 2024. PMID: 38645303 Free PMC article. Review.
Cited by
-
Esperanto for histones: CENP-A, not CenH3, is the centromeric histone H3 variant.Chromosome Res. 2013 Apr;21(2):101-6. doi: 10.1007/s10577-013-9347-y. Epub 2013 Apr 12. Chromosome Res. 2013. PMID: 23580138 Free PMC article.
-
Functions of the centromere and kinetochore in chromosome segregation.Curr Opin Cell Biol. 2013 Jun;25(3):334-40. doi: 10.1016/j.ceb.2013.02.001. Epub 2013 Mar 13. Curr Opin Cell Biol. 2013. PMID: 23490282 Free PMC article. Review.
-
DNA content of a functioning chicken kinetochore.Chromosome Res. 2014 Apr;22(1):7-13. doi: 10.1007/s10577-014-9410-3. Chromosome Res. 2014. PMID: 24633498 Free PMC article.
-
Centromeres of filamentous fungi.Chromosome Res. 2012 Jul;20(5):635-56. doi: 10.1007/s10577-012-9290-3. Chromosome Res. 2012. PMID: 22752455 Free PMC article. Review.
-
ASURA (PHB2) Is Required for Kinetochore Assembly and Subsequent Chromosome Congression.Acta Histochem Cytochem. 2011 Dec 28;44(6):247-58. doi: 10.1267/ahc.11033. Epub 2011 Oct 22. Acta Histochem Cytochem. 2011. PMID: 22282585 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

