Cellular response to influenza virus infection: a potential role for autophagy in CXCL10 and interferon-alpha induction
- PMID: 20473322
- PMCID: PMC4003230
- DOI: 10.1038/cmi.2010.25
Cellular response to influenza virus infection: a potential role for autophagy in CXCL10 and interferon-alpha induction
Abstract
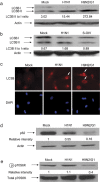
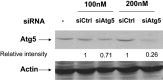
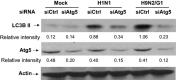
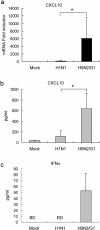
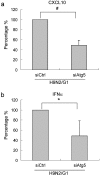
Historically, influenza pandemics have arisen from avian influenza viruses. Avian influenza viruses H5N1 and H9N2 are potential pandemic candidates. Infection of humans with the highly pathogenic avian influenza H5N1 virus is associated with a mortality in excess of 60%, which has been attributed to dysregulation of the cytokine system. Human macrophages and epithelial cells infected with some genotypes of H5N1 and H9N2 viruses express markedly elevated cytokine and chemokine levels when compared with seasonal influenza A subtype H1N1 virus. The mechanisms underlying this cytokine and chemokine hyperinduction are not fully elucidated. In the present study, we demonstrate that autophagy, a tightly regulated homeostatic process for self-digestion of unwanted cellular subcomponents, plays a role in cytokine induction. Autophagy is induced to a greater extent by H9N2/G1, in association with cytokine hyperinduction, compared with H1N1 and the novel pandemic swine-origin influenza A/H1N1 viruses. Using 3-methyladenine to inhibit autophagy and small interfering RNA to silence the autophagy gene, Atg5, we further show that autophagic responses play a role in influenza virus-induced CXCL10 and interferon-alpha expression in primary human blood macrophages. Our results provide new insights into the pathogenic mechanisms of avian influenza viruses.
Figures
Similar articles
-
MXD1 regulates the H9N2 and H1N1 influenza A virus-induced chemokine expression and their replications in human macrophage.J Leukoc Biol. 2020 Nov;108(5):1631-1640. doi: 10.1002/JLB.4MA0620-703RR. Epub 2020 Aug 13. J Leukoc Biol. 2020. PMID: 32794336
-
Mammalian innate resistance to highly pathogenic avian influenza H5N1 virus infection is mediated through reduced proinflammation and infectious virus release.J Virol. 2012 Sep;86(17):9201-10. doi: 10.1128/JVI.00244-12. Epub 2012 Jun 20. J Virol. 2012. PMID: 22718824 Free PMC article.
-
Differential expression of chemokines and their receptors in adult and neonatal macrophages infected with human or avian influenza viruses.J Infect Dis. 2006 Jul 1;194(1):61-70. doi: 10.1086/504690. Epub 2006 May 26. J Infect Dis. 2006. PMID: 16741883 Free PMC article.
-
[Swine influenza virus: evolution mechanism and epidemic characterization--a review].Wei Sheng Wu Xue Bao. 2009 Sep;49(9):1138-45. Wei Sheng Wu Xue Bao. 2009. PMID: 20030049 Review. Chinese.
-
The use of nonhuman primates in research on seasonal, pandemic and avian influenza, 1893-2014.Antiviral Res. 2015 May;117:75-98. doi: 10.1016/j.antiviral.2015.02.011. Epub 2015 Mar 5. Antiviral Res. 2015. PMID: 25746173 Free PMC article. Review.
Cited by
-
Potential Molecular Mechanisms and Remdesivir Treatment for Acute Respiratory Syndrome Corona Virus 2 Infection/COVID 19 Through RNA Sequencing and Bioinformatics Analysis.Bioinform Biol Insights. 2021 Dec 23;15:11779322211067365. doi: 10.1177/11779322211067365. eCollection 2021. Bioinform Biol Insights. 2021. PMID: 34992355 Free PMC article.
-
Autophagy: The multi-purpose bridge in viral infections and host cells.Rev Med Virol. 2018 Jul;28(4):e1973. doi: 10.1002/rmv.1973. Epub 2018 Apr 30. Rev Med Virol. 2018. PMID: 29709097 Free PMC article. Review.
-
Coronaviruses construct an interconnection way with ERAD and autophagy.Future Microbiol. 2021 Sep;16(14):1135-1151. doi: 10.2217/fmb-2021-0044. Epub 2021 Sep 1. Future Microbiol. 2021. PMID: 34468179 Free PMC article. Review.
-
Autophagy in Multiple Sclerosis: Two Sides of the Same Coin.Front Cell Neurosci. 2020 Nov 20;14:603710. doi: 10.3389/fncel.2020.603710. eCollection 2020. Front Cell Neurosci. 2020. PMID: 33328897 Free PMC article. Review.
-
Bioinformatics analyses of significant genes, related pathways, and candidate diagnostic biomarkers and molecular targets in SARS-CoV-2/COVID-19.Gene Rep. 2020 Dec;21:100956. doi: 10.1016/j.genrep.2020.100956. Epub 2020 Nov 4. Gene Rep. 2020. PMID: 33553808 Free PMC article.
References
-
- Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, et al. Human infection with influenza H9N2. Lancet. 1999;354:916–917. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

