Phylodynamic reconstruction reveals norovirus GII.4 epidemic expansions and their molecular determinants
- PMID: 20463813
- PMCID: PMC2865530
- DOI: 10.1371/journal.ppat.1000884
Phylodynamic reconstruction reveals norovirus GII.4 epidemic expansions and their molecular determinants
Abstract
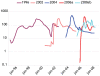
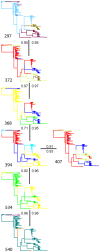
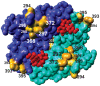
Noroviruses are the most common cause of viral gastroenteritis. An increase in the number of globally reported norovirus outbreaks was seen the past decade, especially for outbreaks caused by successive genogroup II genotype 4 (GII.4) variants. Whether this observed increase was due to an upswing in the number of infections, or to a surveillance artifact caused by heightened awareness and concomitant improved reporting, remained unclear. Therefore, we set out to study the population structure and changes thereof of GII.4 strains detected through systematic outbreak surveillance since the early 1990s. We collected 1383 partial polymerase and 194 full capsid GII.4 sequences. A Bayesian MCMC coalescent analysis revealed an increase in the number of GII.4 infections during the last decade. The GII.4 strains included in our analyses evolved at a rate of 4.3-9.0x10(-3) mutations per site per year, and share a most recent common ancestor in the early 1980s. Determinants of adaptation in the capsid protein were studied using different maximum likelihood approaches to identify sites subject to diversifying or directional selection and sites that co-evolved. While a number of the computationally determined adaptively evolving sites were on the surface of the capsid and possible subject to immune selection, we also detected sites that were subject to constrained or compensatory evolution due to secondary RNA structures, relevant in virus-replication. We highlight codons that may prove useful in identifying emerging novel variants, and, using these, indicate that the novel 2008 variant is more likely to cause a future epidemic than the 2007 variant. While norovirus infections are generally mild and self-limiting, more severe outcomes of infection frequently occur in elderly and immunocompromized people, and no treatment is available. The observed pattern of continually emerging novel variants of GII.4, causing elevated numbers of infections, is therefore a cause for concern.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Norovirus GII.4 and GII.7 capsid sequences undergo positive selection in chronically infected patients.Infect Genet Evol. 2012 Mar;12(2):461-6. doi: 10.1016/j.meegid.2012.01.020. Epub 2012 Jan 30. Infect Genet Evol. 2012. PMID: 22310302
-
Genetic analyses of GII.17 norovirus strains in diarrheal disease outbreaks from December 2014 to March 2015 in Japan reveal a novel polymerase sequence and amino acid substitutions in the capsid region.Euro Surveill. 2015 Jul 2;20(26):21173. doi: 10.2807/1560-7917.es2015.20.26.21173. Euro Surveill. 2015. PMID: 26159307
-
Comparative evolution of GII.3 and GII.4 norovirus over a 31-year period.J Virol. 2011 Sep;85(17):8656-66. doi: 10.1128/JVI.00472-11. Epub 2011 Jun 29. J Virol. 2011. PMID: 21715504 Free PMC article.
-
Rapid evolution of pandemic noroviruses of the GII.4 lineage.PLoS Pathog. 2010 Mar 26;6(3):e1000831. doi: 10.1371/journal.ppat.1000831. PLoS Pathog. 2010. PMID: 20360972 Free PMC article.
-
Molecular and Genetics-Based Systems for Tracing the Evolution and Exploring the Mechanisms of Human Norovirus Infections.Int J Mol Sci. 2023 May 22;24(10):9093. doi: 10.3390/ijms24109093. Int J Mol Sci. 2023. PMID: 37240438 Free PMC article. Review.
Cited by
-
Strain-dependent norovirus bioaccumulation in oysters.Appl Environ Microbiol. 2011 May;77(10):3189-96. doi: 10.1128/AEM.03010-10. Epub 2011 Mar 25. Appl Environ Microbiol. 2011. PMID: 21441327 Free PMC article.
-
Temporal Evolutionary Dynamics of Norovirus GII.4 Variants in China between 2004 and 2015.PLoS One. 2016 Sep 20;11(9):e0163166. doi: 10.1371/journal.pone.0163166. eCollection 2016. PLoS One. 2016. PMID: 27649572 Free PMC article.
-
Caliciviruses in hospitalized children, São Luís, Maranhão, 1997-1999: detection of norovirus GII.12.Braz J Microbiol. 2016 Jul-Sep;47(3):724-30. doi: 10.1016/j.bjm.2016.04.008. Epub 2016 Apr 22. Braz J Microbiol. 2016. PMID: 27161199 Free PMC article.
-
Identification of Human Norovirus GII.3 Blockade Antibody Epitopes.Viruses. 2021 Oct 13;13(10):2058. doi: 10.3390/v13102058. Viruses. 2021. PMID: 34696487 Free PMC article.
-
Inference for nonlinear epidemiological models using genealogies and time series.PLoS Comput Biol. 2011 Aug;7(8):e1002136. doi: 10.1371/journal.pcbi.1002136. Epub 2011 Aug 25. PLoS Comput Biol. 2011. PMID: 21901082 Free PMC article.
References
-
- Atmar RL, Estes MK. The epidemiologic and clinical importance of norovirus infection. Gastroenterol Clin North Am. 2006;35:275–90, viii. - PubMed
-
- Patel MM, Hall AJ, Vinje J, Parashar UD. Noroviruses: a comprehensive review. J Clin Virol. 2009;44:1–8. - PubMed
-
- Rockx B, De Wit M, Vennema H, Vinje J, De Bruin E, et al. Natural history of human calicivirus infection: a prospective cohort study. Clin Infect Dis. 2002;35:246–53. - PubMed
-
- de Wit MA, Koopmans MP, Kortbeek LM, Wannet WJ, Vinje J, et al. Sensor, a population-based cohort study on gastroenteritis in the Netherlands: incidence and etiology. Am J Epidemiol. 2001;154:666–74. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

