Genome-wide estimation of firing efficiencies of origins of DNA replication from time-course copy number variation data
- PMID: 20462459
- PMCID: PMC2885374
- DOI: 10.1186/1471-2105-11-247
Genome-wide estimation of firing efficiencies of origins of DNA replication from time-course copy number variation data
Abstract
Background: DNA replication is a fundamental biological process during S phase of cell division. It is initiated from several hundreds of origins along whole chromosome with different firing efficiencies (or frequency of usage). Direct measurement of origin firing efficiency by techniques such as DNA combing are time-consuming and lack the ability to measure all origins. Recent genome-wide study of DNA replication approximated origin firing efficiency by indirectly measuring other quantities related to replication. However, these approximation methods do not reflect properties of origin firing and may lead to inappropriate estimations.
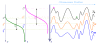
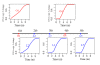
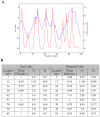
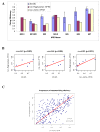
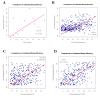
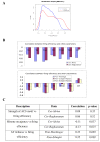
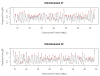
Results: In this paper, we develop a probabilistic model - Spanned Firing Time Model (SFTM) to characterize DNA replication process. The proposed model reflects current understandings about DNA replication. Origins in an individual cell may initiate replication randomly within a time window, but the population average exhibits a temporal program with some origins replicated early and the others late. By estimating DNA origin firing time and fork moving velocity from genome-wide time-course S-phase copy number variation data, we could estimate firing efficiency of all origins. The estimated firing efficiency is correlated well with the previous studies in fission and budding yeasts.
Conclusions: The new probabilistic model enables sensitive identification of origins as well as genome-wide estimation of origin firing efficiency. We have successfully estimated firing efficiencies of all origins in S. cerevisiae, S. pombe and human chromosomes 21 and 22.
Figures
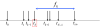
Similar articles
-
DNA replication origins fire stochastically in fission yeast.Mol Biol Cell. 2006 Jan;17(1):308-16. doi: 10.1091/mbc.e05-07-0657. Epub 2005 Oct 26. Mol Biol Cell. 2006. PMID: 16251353 Free PMC article.
-
A variable fork rate affects timing of origin firing and S phase dynamics in Saccharomyces cerevisiae.J Biotechnol. 2013 Oct 20;168(2):174-84. doi: 10.1016/j.jbiotec.2013.06.022. Epub 2013 Jul 9. J Biotechnol. 2013. PMID: 23850861
-
Genome-wide characterization of fission yeast DNA replication origins.EMBO J. 2006 Nov 1;25(21):5171-9. doi: 10.1038/sj.emboj.7601390. Epub 2006 Oct 19. EMBO J. 2006. PMID: 17053780 Free PMC article.
-
Genomic specification and epigenetic regulation of eukaryotic DNA replication origins.EMBO J. 2004 Nov 10;23(22):4365-70. doi: 10.1038/sj.emboj.7600450. Epub 2004 Oct 28. EMBO J. 2004. PMID: 15510221 Free PMC article. Review.
-
DNA replication in the fission yeast: robustness in the face of uncertainty.Yeast. 2006 Oct 15;23(13):951-62. doi: 10.1002/yea.1416. Yeast. 2006. PMID: 17072888 Review.
Cited by
-
DNA replication timing.Cold Spring Harb Perspect Biol. 2013 Aug 1;5(8):a010132. doi: 10.1101/cshperspect.a010132. Cold Spring Harb Perspect Biol. 2013. PMID: 23838440 Free PMC article. Review.
-
Mathematical modeling of genome replication.Phys Rev E Stat Nonlin Soft Matter Phys. 2012 Sep;86(3 Pt 1):031916. doi: 10.1103/PhysRevE.86.031916. Epub 2012 Sep 17. Phys Rev E Stat Nonlin Soft Matter Phys. 2012. PMID: 23030953 Free PMC article.
-
Dynamics of DNA replication in yeast.Phys Rev Lett. 2011 Aug 5;107(6):068103. doi: 10.1103/PhysRevLett.107.068103. Epub 2011 Aug 4. Phys Rev Lett. 2011. PMID: 21902372 Free PMC article.
-
The spatiotemporal program of replication in the genome of Lachancea kluyveri.Genome Biol Evol. 2013;5(2):370-88. doi: 10.1093/gbe/evt014. Genome Biol Evol. 2013. PMID: 23355306 Free PMC article.
-
Replication timing and its emergence from stochastic processes.Trends Genet. 2012 Aug;28(8):374-81. doi: 10.1016/j.tig.2012.03.011. Epub 2012 Apr 18. Trends Genet. 2012. PMID: 22520729 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

