Non-canonical activation of Notch signaling/target genes in vertebrates
- PMID: 20458516
- PMCID: PMC11115867
- DOI: 10.1007/s00018-010-0391-x
Non-canonical activation of Notch signaling/target genes in vertebrates
Abstract
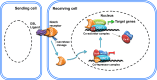
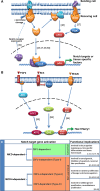
Evolutionarily conserved Notch signaling orchestrates diverse physiological mechanisms during metazoan development and homeostasis. Classically, ligand-activated Notch receptors transduce the signaling cascade through the interaction of DNA-bound CBF1-co-repressor complex. However, recent reports have demonstrated execution of a CBF1-independent Notch pathway through signaling cross-talks in various cells/tissues. Here, we have tried to congregate the reports that describe the non-canonical/CBF1-independent Notch signaling and target gene activation in vertebrates with specific emphasis on their functional relevance.
Figures
Similar articles
-
RBPJ/CBF1 interacts with L3MBTL3/MBT1 to promote repression of Notch signaling via histone demethylase KDM1A/LSD1.EMBO J. 2017 Nov 2;36(21):3232-3249. doi: 10.15252/embj.201796525. Epub 2017 Oct 13. EMBO J. 2017. PMID: 29030483 Free PMC article.
-
A bright single-cell resolution live imaging reporter of Notch signaling in the mouse.BMC Dev Biol. 2013 Apr 25;13:15. doi: 10.1186/1471-213X-13-15. BMC Dev Biol. 2013. PMID: 23617465 Free PMC article.
-
ATF2 maintains a subset of neural progenitors through CBF1/Notch independent Hes-1 expression and synergistically activates the expression of Hes-1 in Notch-dependent neural progenitors.J Neurochem. 2010 May;113(4):807-18. doi: 10.1111/j.1471-4159.2010.06574.x. Epub 2010 Jan 8. J Neurochem. 2010. PMID: 20067572
-
Notch-independent functions of CSL.Curr Top Dev Biol. 2011;97:55-74. doi: 10.1016/B978-0-12-385975-4.00009-7. Curr Top Dev Biol. 2011. PMID: 22074602 Review.
-
The Notch signaling pathway: transcriptional regulation at Notch target genes.Cell Mol Life Sci. 2009 May;66(10):1631-46. doi: 10.1007/s00018-009-8668-7. Cell Mol Life Sci. 2009. PMID: 19165418 Free PMC article. Review.
Cited by
-
C-terminal deletion of NOTCH1 intracellular domain (N1ICD) increases its stability but does not amplify and recapitulate N1ICD-dependent signalling.Sci Rep. 2017 Jul 11;7(1):5034. doi: 10.1038/s41598-017-05119-0. Sci Rep. 2017. PMID: 28698562 Free PMC article.
-
Hes1: the maestro in neurogenesis.Cell Mol Life Sci. 2016 Nov;73(21):4019-42. doi: 10.1007/s00018-016-2277-z. Epub 2016 May 27. Cell Mol Life Sci. 2016. PMID: 27233500 Free PMC article. Review.
-
Localization of sequences in a protein (ORF2) encoded by the latency-related gene of bovine herpesvirus 1 that inhibits apoptosis and interferes with Notch1-mediated trans-activation of the bICP0 promoter.J Virol. 2011 Dec;85(23):12124-33. doi: 10.1128/JVI.05478-11. Epub 2011 Sep 21. J Virol. 2011. PMID: 21937659 Free PMC article.
-
Canonical Notch signaling is dispensable for adult steady-state and stress myelo-erythropoiesis.Blood. 2018 Apr 12;131(15):1712-1719. doi: 10.1182/blood-2017-06-788505. Epub 2018 Jan 16. Blood. 2018. PMID: 29339402 Free PMC article.
-
Notch signaling pathway in cancer: from mechanistic insights to targeted therapies.Signal Transduct Target Ther. 2024 May 27;9(1):128. doi: 10.1038/s41392-024-01828-x. Signal Transduct Target Ther. 2024. PMID: 38797752 Free PMC article. Review.
References
-
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. - PubMed
-
- Mumm JS, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228:151–165. - PubMed
-
- Honjo T. The shortest path from the surface to the nucleus: RBP-J kappa/Su(H) transcription factor. Genes Cells. 1996;1:1–9. - PubMed
-
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

