Alternative tasks of Drosophila tan in neurotransmitter recycling versus cuticle sclerotization disclosed by kinetic properties
- PMID: 20439462
- PMCID: PMC2898368
- DOI: 10.1074/jbc.M110.120170
Alternative tasks of Drosophila tan in neurotransmitter recycling versus cuticle sclerotization disclosed by kinetic properties
Abstract
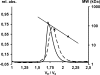
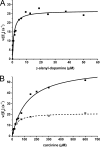
Upon a stimulus of light, histamine is released from Drosophila photoreceptor axonal endings. It is taken up into glia where Ebony converts it into beta-alanyl-histamine (carcinine). Carcinine moves into photoreceptor cells and is there cleaved into beta-alanine and histamine by Tan activity. Tan thus provides a key function in the recycling pathway of the neurotransmitter histamine. It is also involved in the process of cuticle formation. There, it cleaves beta-alanyl-dopamine, a major component in cuticle sclerotization. Active Tan enzyme is generated by a self-processing proteolytic cleavage from a pre-protein at a conserved Gly-Cys sequence motif. We confirmed the dependence on the Gly-Cys motif by in vitro mutagenesis. Processing time delays the rise to full Tan activity up to 3 h behind its putative circadian RNA expression in head. To investigate its pleiotropic functions, we have expressed Tan as a His(6) fusion protein in Escherichia coli and have purified it to homogeneity. We found wild type and mutant His(6)-Tan protein co-migrating in size exclusion chromatography with a molecular weight compatible with homodimer formation. We conclude that dimer formation is preceding pre-protein processing. Drosophila tan(1) null mutant analysis revealed that amino acid Arg(217) is absolutely required for processing. Substitution of Met(256) in tan(5), on the contrary, does not affect processing extensively but renders it prone to degradation. This also leads to a strong tan phenotype although His(6)-Tan(5) retains activity. Kinetic parameters of Tan reveal characteristic differences in K(m) and k(cat) values of carcinine and beta-alanyl-dopamine cleavage, which conclusively illustrate the divergent tasks met by Tan.
Figures
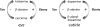




Similar articles
-
tan and ebony genes regulate a novel pathway for transmitter metabolism at fly photoreceptor terminals.J Neurosci. 2002 Dec 15;22(24):10549-57. doi: 10.1523/JNEUROSCI.22-24-10549.2002. J Neurosci. 2002. PMID: 12486147 Free PMC article.
-
The role of carcinine in signaling at the Drosophila photoreceptor synapse.PLoS Genet. 2007 Dec;3(12):e206. doi: 10.1371/journal.pgen.0030206. PLoS Genet. 2007. PMID: 18069895 Free PMC article.
-
Quantification of Histamine and Carcinine in Drosophila melanogaster Tissues.ACS Chem Neurosci. 2016 Mar 16;7(3):407-14. doi: 10.1021/acschemneuro.5b00326. Epub 2016 Jan 28. ACS Chem Neurosci. 2016. PMID: 26765065 Free PMC article.
-
Pigmentation and behavior: potential association through pleiotropic genes in Drosophila.Genes Genet Syst. 2013;88(3):165-74. doi: 10.1266/ggs.88.165. Genes Genet Syst. 2013. PMID: 24025245 Review.
-
Chromatin mechanisms in Drosophila dosage compensation.Prog Mol Subcell Biol. 2005;38:123-49. doi: 10.1007/3-540-27310-7_5. Prog Mol Subcell Biol. 2005. PMID: 15881893 Review.
Cited by
-
Catecholamine Derivatives as Novel Crosslinkers for the Synthesis of Versatile Biopolymers.J Funct Biomater. 2023 Sep 1;14(9):449. doi: 10.3390/jfb14090449. J Funct Biomater. 2023. PMID: 37754863 Free PMC article. Review.
-
Combinatorial expression of ebony and tan generates body color variation from nymph through adult stages in the cricket, Gryllus bimaculatus.PLoS One. 2023 May 18;18(5):e0285934. doi: 10.1371/journal.pone.0285934. eCollection 2023. PLoS One. 2023. PMID: 37200362 Free PMC article.
-
Strong epistatic and additive effects of linked candidate SNPs for Drosophila pigmentation have implications for analysis of genome-wide association studies results.Genome Biol. 2017 Jul 3;18(1):126. doi: 10.1186/s13059-017-1262-7. Genome Biol. 2017. PMID: 28673357 Free PMC article.
-
Critical Analysis of the Melanogenic Pathway in Insects and Higher Animals.Int J Mol Sci. 2016 Oct 20;17(10):1753. doi: 10.3390/ijms17101753. Int J Mol Sci. 2016. PMID: 27775611 Free PMC article. Review.
-
The metabolism of histamine in the Drosophila optic lobe involves an ommatidial pathway: β-alanine recycles through the retina.J Exp Biol. 2012 Apr 15;215(Pt 8):1399-411. doi: 10.1242/jeb.060699. J Exp Biol. 2012. PMID: 22442379 Free PMC article.
References
-
- Stuart A. E., Borycz J., Meinertzhagen I. A. (2007) Prog. Neurobiol. 82, 202–227 - PubMed
-
- Richardt A., Kemme T., Wagner S., Schwarzer D., Marahiel M. A., Hovemann B. T. (2003) J. Biol. Chem. 278, 41160–41166 - PubMed
-
- Wagner S., Heseding C., Szlachta K., True J. R., Prinz H., Hovemann B. T. (2007) J. Comp. Neurol. 500, 601–611 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

