The molecular evolution of PL10 homologs
- PMID: 20438638
- PMCID: PMC2874800
- DOI: 10.1186/1471-2148-10-127
The molecular evolution of PL10 homologs
Abstract
Background: PL10 homologs exist in a wide range of eukaryotes from yeast, plants to animals. They share a DEAD motif and belong to the DEAD-box polypeptide 3 (DDX3) subfamily with a major role in RNA metabolism. The lineage-specific expression patterns and various genomic structures and locations of PL10 homologs indicate these homologs have an interesting evolutionary history.
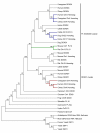
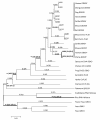
Results: Phylogenetic analyses revealed that, in addition to the sex chromosome-linked PL10 homologs, DDX3X and DDX3Y, a single autosomal PL10 putative homologous sequence is present in each genome of the studied non-rodent eutheria. These autosomal homologous sequences originated from the retroposition of DDX3X but were pseudogenized during the evolution. In rodents, besides Ddx3x and Ddx3y, we found not only Pl10 but another autosomal homologous region, both of which also originated from the Ddx3x retroposition. These retropositions occurred after the divergence of eutheria and opossum. In contrast, an additional X putative homologous sequence was detected in primates and originated from the transposition of DDX3Y. The evolution of PL10 homologs was under positive selection and the elevated Ka/Ks ratios were observed in the eutherian lineages for DDX3Y but not PL10 and DDX3X, suggesting relaxed selective constraints on DDX3Y. Contrary to the highly conserved domains, several sites with relaxed selective constraints flanking the domains in the mammalian PL10 homologs may play roles in enhancing the gene function in a lineage-specific manner.
Conclusion: The eutherian DDX3X/DDX3Y in the X/Y-added region originated from the translocation of the ancient PL10 ortholog on the ancestral autosome, whereas the eutherian PL10 was retroposed from DDX3X. In addition to the functional PL10/DDX3X/DDX3Y, conserved homologous regions on the autosomes and X chromosome are present. The autosomal homologs were also derived from DDX3X, whereas the additional X-homologs were derived from DDX3Y. These homologs were apparently pseudogenized but may still be active transcriptionally. The evolution of PL10 homologs was positively selected.
Figures


Similar articles
-
Molecular characterization of the DDX3Y gene and its homologs in cattle.Cytogenet Genome Res. 2009;126(4):318-28. doi: 10.1159/000266168. Epub 2009 Dec 11. Cytogenet Genome Res. 2009. PMID: 20016128
-
DDX3X, the X homologue of AZFa gene DDX3Y, expresses a complex pattern of transcript variants only in the male germ line.Mol Hum Reprod. 2014 Dec;20(12):1208-22. doi: 10.1093/molehr/gau081. Epub 2014 Sep 10. Mol Hum Reprod. 2014. PMID: 25208899
-
Human DDX3Y, the Y-encoded isoform of RNA helicase DDX3, rescues a hamster temperature-sensitive ET24 mutant cell line with a DDX3X mutation.Exp Cell Res. 2004 Oct 15;300(1):213-22. doi: 10.1016/j.yexcr.2004.07.005. Exp Cell Res. 2004. PMID: 15383328
-
The DDX3 subfamily of the DEAD box helicases: divergent roles as unveiled by studying different organisms and in vitro assays.Curr Med Chem. 2007;14(23):2517-25. doi: 10.2174/092986707782023677. Curr Med Chem. 2007. PMID: 17979704 Review.
-
The variant landscape and function of DDX3X in cancer and neurodevelopmental disorders.Trends Mol Med. 2023 Sep;29(9):726-739. doi: 10.1016/j.molmed.2023.06.003. Epub 2023 Jul 6. Trends Mol Med. 2023. PMID: 37422363 Review.
Cited by
-
The RNA Helicase Ded1 from Yeast Is Associated with the Signal Recognition Particle and Is Regulated by SRP21.Molecules. 2024 Jun 20;29(12):2944. doi: 10.3390/molecules29122944. Molecules. 2024. PMID: 38931009 Free PMC article.
-
mRNA helicases: the tacticians of translational control.Nat Rev Mol Cell Biol. 2011 Apr;12(4):235-45. doi: 10.1038/nrm3083. Nat Rev Mol Cell Biol. 2011. PMID: 21427765 Review.
-
Comparative Evolution of Duplicated Ddx3 Genes in Teleosts: Insights from Japanese Flounder, Paralichthys olivaceus.G3 (Bethesda). 2015 Jun 24;5(8):1765-73. doi: 10.1534/g3.115.018911. G3 (Bethesda). 2015. PMID: 26109358 Free PMC article.
-
Targeting RNA helicase DDX3 in stem cell maintenance and teratoma formation.Genes Cancer. 2019 Feb;10(1-2):11-20. doi: 10.18632/genesandcancer.187. Genes Cancer. 2019. PMID: 30899416 Free PMC article.
-
Mechanistic characterization of the DEAD-box RNA helicase Ded1 from yeast as revealed by a novel technique using single-molecule magnetic tweezers.Nucleic Acids Res. 2019 Apr 23;47(7):3699-3710. doi: 10.1093/nar/gkz057. Nucleic Acids Res. 2019. PMID: 30993346 Free PMC article.
References
-
- Leroy P, Seboun E, Mattei MG, Fellous M, Bishop CE. Testis-specific transcripts detected by a human Y-DNA-derived probe. Development. 1987;101(Suppl):177–183. - PubMed
-
- UniProt. http://www.uniprot.org/
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

