Clinical assessment incorporating a personal genome
- PMID: 20435227
- PMCID: PMC2937184
- DOI: 10.1016/S0140-6736(10)60452-7
Clinical assessment incorporating a personal genome
Abstract
Background: The cost of genomic information has fallen steeply, but the clinical translation of genetic risk estimates remains unclear. We aimed to undertake an integrated analysis of a complete human genome in a clinical context.
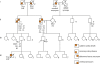
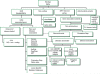
Methods: We assessed a patient with a family history of vascular disease and early sudden death. Clinical assessment included analysis of this patient's full genome sequence, risk prediction for coronary artery disease, screening for causes of sudden cardiac death, and genetic counselling. Genetic analysis included the development of novel methods for the integration of whole genome and clinical risk. Disease and risk analysis focused on prediction of genetic risk of variants associated with mendelian disease, recognised drug responses, and pathogenicity for novel variants. We queried disease-specific mutation databases and pharmacogenomics databases to identify genes and mutations with known associations with disease and drug response. We estimated post-test probabilities of disease by applying likelihood ratios derived from integration of multiple common variants to age-appropriate and sex-appropriate pre-test probabilities. We also accounted for gene-environment interactions and conditionally dependent risks.
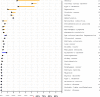
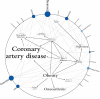
Findings: Analysis of 2.6 million single nucleotide polymorphisms and 752 copy number variations showed increased genetic risk for myocardial infarction, type 2 diabetes, and some cancers. We discovered rare variants in three genes that are clinically associated with sudden cardiac death-TMEM43, DSP, and MYBPC3. A variant in LPA was consistent with a family history of coronary artery disease. The patient had a heterozygous null mutation in CYP2C19 suggesting probable clopidogrel resistance, several variants associated with a positive response to lipid-lowering therapy, and variants in CYP4F2 and VKORC1 that suggest he might have a low initial dosing requirement for warfarin. Many variants of uncertain importance were reported.
Interpretation: Although challenges remain, our results suggest that whole-genome sequencing can yield useful and clinically relevant information for individual patients.
Funding: National Institute of General Medical Sciences; National Heart, Lung And Blood Institute; National Human Genome Research Institute; Howard Hughes Medical Institute; National Library of Medicine, Lucile Packard Foundation for Children's Health; Hewlett Packard Foundation; Breetwor Family Foundation.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures

Comment in
-
The personal genome--the future of personalised medicine?Lancet. 2010 May 1;375(9725):1497-8. doi: 10.1016/S0140-6736(10)60598-3. Lancet. 2010. PMID: 20435212 No abstract available.
-
Clinical assessment incorporating a personal genome.Lancet. 2010 Sep 11;376(9744):869; author reply 869-70. doi: 10.1016/S0140-6736(10)61404-3. Lancet. 2010. PMID: 20833292 No abstract available.
Similar articles
-
A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose.PLoS Genet. 2009 Mar;5(3):e1000433. doi: 10.1371/journal.pgen.1000433. Epub 2009 Mar 20. PLoS Genet. 2009. PMID: 19300499 Free PMC article.
-
An evaluation of nine genetic variants related to metabolism and mechanism of action of warfarin as applied to stable dose prediction.J Thromb Thrombolysis. 2010 Oct;30(3):358-64. doi: 10.1007/s11239-010-0467-3. J Thromb Thrombolysis. 2010. PMID: 20499136
-
Dependency of phenprocoumon dosage on polymorphisms in the VKORC1, CYP2C9, and CYP4F2 genes.Pharmacogenet Genomics. 2011 Jan;21(1):26-34. doi: 10.1097/FPC.0b013e32834154fb. Pharmacogenet Genomics. 2011. PMID: 21063236
-
Clinical application of cardiovascular pharmacogenetics.J Am Coll Cardiol. 2012 Jul 3;60(1):9-20. doi: 10.1016/j.jacc.2012.01.067. J Am Coll Cardiol. 2012. PMID: 22742397 Review.
-
Emerging genomic applications in coronary artery disease.JACC Cardiovasc Interv. 2011 May;4(5):473-82. doi: 10.1016/j.jcin.2010.12.016. JACC Cardiovasc Interv. 2011. PMID: 21596318 Free PMC article. Review.
Cited by
-
Workflow for the Implementation of Precision Genomics in Healthcare.Front Genet. 2020 Jun 30;11:619. doi: 10.3389/fgene.2020.00619. eCollection 2020. Front Genet. 2020. PMID: 32695137 Free PMC article.
-
Evidence for Clinical Implementation of Pharmacogenomics in Cardiac Drugs.Mayo Clin Proc. 2015 Jun;90(6):716-29. doi: 10.1016/j.mayocp.2015.03.016. Mayo Clin Proc. 2015. PMID: 26046407 Free PMC article.
-
Public attitudes towards genetic testing revisited: comparing opinions between 2002 and 2010.Eur J Hum Genet. 2013 Aug;21(8):793-9. doi: 10.1038/ejhg.2012.271. Epub 2012 Dec 19. Eur J Hum Genet. 2013. PMID: 23249955 Free PMC article.
-
Deletions of recessive disease genes: CNV contribution to carrier states and disease-causing alleles.Genome Res. 2013 Sep;23(9):1383-94. doi: 10.1101/gr.156075.113. Epub 2013 May 16. Genome Res. 2013. PMID: 23685542 Free PMC article.
-
A microfluidic device for preparing next generation DNA sequencing libraries and for automating other laboratory protocols that require one or more column chromatography steps.PLoS One. 2013 Jul 24;8(7):e64084. doi: 10.1371/journal.pone.0064084. Print 2013. PLoS One. 2013. PMID: 23894273 Free PMC article.
References
-
- Wheeler DA, Srinivasan M, Egholm M, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452(7189):872–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R24 GM061374/GM/NIGMS NIH HHS/United States
- T15 LM007033-23/LM/NLM NIH HHS/United States
- K08 HL083914-04/HL/NHLBI NIH HHS/United States
- F32HL097462/HL/NHLBI NIH HHS/United States
- GM61374/GM/NIGMS NIH HHS/United States
- U01 GM061374/GM/NIGMS NIH HHS/United States
- R01 GM079719-04/GM/NIGMS NIH HHS/United States
- R01 GM079719/GM/NIGMS NIH HHS/United States
- F32 HL097462-01/HL/NHLBI NIH HHS/United States
- GM079719/GM/NIGMS NIH HHS/United States
- U01 GM061374-07/GM/NIGMS NIH HHS/United States
- HG003389/HG/NHGRI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 LM009719-02/LM/NLM NIH HHS/United States
- K08 HL083914/HL/NHLBI NIH HHS/United States
- T15 LM007033/LM/NLM NIH HHS/United States
- F32 HL097462/HL/NHLBI NIH HHS/United States
- P50 HG003389/HG/NHGRI NIH HHS/United States
- T32 HL094274/HL/NHLBI NIH HHS/United States
- P50 HG003389-05/HG/NHGRI NIH HHS/United States
- LM009719/LM/NLM NIH HHS/United States
- R01 LM009719/LM/NLM NIH HHS/United States
- LM007033/LM/NLM NIH HHS/United States
- DP2 OD006511/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

