Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction
- PMID: 20410271
- PMCID: PMC2903277
- DOI: 10.1128/JVI.01519-09
Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction
Abstract
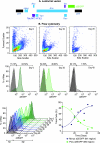
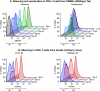
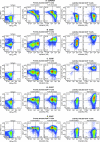
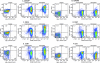
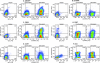
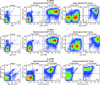
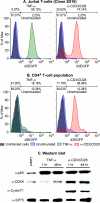
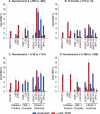
The development of suitable experimental systems for studying HIV latency in primary cells that permit detailed biochemical analyses and the screening of drugs is a critical step in the effort to develop viral eradication strategies. Primary CD4(+) T cells were isolated from peripheral blood and amplified by antibodies to the T-cell receptor (TCR). The cells were then infected by lentiviral vectors carrying fluorescent reporters and either the wild-type Tat gene or the attenuated H13L Tat gene. After sorting for the positive cells and reamplification, the infected cells were allowed to spontaneously enter latency by long-term cultivation on the H80 feeder cell line in the absence of TCR stimulation. By 6 weeks almost all of the cells lost fluorescent protein marker expression; however, more than 95% of these latently infected cells could be reactivated after stimulation of the TCR by alpha-CD3/CD28 antibodies. Chromatin immunoprecipitation assays showed that, analogously to Jurkat T cells, latent proviruses in primary CD4(+) T cells are enriched in heterochromatic markers, including high levels of CBF-1, histone deacetylases, and methylated histones. Upon TCR activation, there was recruitment of NF-kappaB to the promoter and conversion of heterochromatin structures present on the latent provirus to active euchromatin structures containing acetylated histones. Surprisingly, latently infected primary cells cannot be induced by tumor necrosis factor alpha because of a restriction in P-TEFb levels, which can be overcome by activation of the TCR. Thus, a combination of restrictive chromatin structures at the HIV long terminal repeat and limiting P-TEFb levels contribute to transcriptional silencing leading to latency in primary CD4(+) T cells.
Figures

Similar articles
-
T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway.J Mol Biol. 2011 Jul 29;410(5):896-916. doi: 10.1016/j.jmb.2011.03.054. J Mol Biol. 2011. PMID: 21763495 Free PMC article.
-
Efficient Non-Epigenetic Activation of HIV Latency through the T-Cell Receptor Signalosome.Viruses. 2020 Aug 8;12(8):868. doi: 10.3390/v12080868. Viruses. 2020. PMID: 32784426 Free PMC article. Review.
-
Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency.J Virol. 2008 Dec;82(24):12291-303. doi: 10.1128/JVI.01383-08. Epub 2008 Oct 1. J Virol. 2008. PMID: 18829756 Free PMC article.
-
Biogenesis of P-TEFb in CD4+ T cells to reverse HIV latency is mediated by protein kinase C (PKC)-independent signaling pathways.PLoS Pathog. 2021 Sep 16;17(9):e1009581. doi: 10.1371/journal.ppat.1009581. eCollection 2021 Sep. PLoS Pathog. 2021. PMID: 34529720 Free PMC article.
-
The cell biology of HIV-1 latency and rebound.Retrovirology. 2024 Apr 5;21(1):6. doi: 10.1186/s12977-024-00639-w. Retrovirology. 2024. PMID: 38580979 Free PMC article. Review.
Cited by
-
Targeting Epigenetics to Cure HIV-1: Lessons From (and for) Cancer Treatment.Front Cell Infect Microbiol. 2021 May 7;11:668637. doi: 10.3389/fcimb.2021.668637. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34026665 Free PMC article. Review.
-
Targeted Chromatinization and Repression of HIV-1 Provirus Transcription with Repurposed CRISPR/Cas9.Viruses. 2020 Oct 12;12(10):1154. doi: 10.3390/v12101154. Viruses. 2020. PMID: 33053801 Free PMC article.
-
The immunological synapse: the gateway to the HIV reservoir.Immunol Rev. 2013 Jul;254(1):305-25. doi: 10.1111/imr.12080. Immunol Rev. 2013. PMID: 23772628 Free PMC article. Review.
-
Targeting HIV latency: pharmacologic strategies toward eradication.Drug Discov Today. 2013 Jun;18(11-12):541-51. doi: 10.1016/j.drudis.2012.12.008. Epub 2012 Dec 25. Drug Discov Today. 2013. PMID: 23270785 Free PMC article. Review.
-
Towards an HIV cure: a global scientific strategy.Nat Rev Immunol. 2012 Jul 20;12(8):607-14. doi: 10.1038/nri3262. Nat Rev Immunol. 2012. PMID: 22814509 Free PMC article. Review.
References
-
- Biglione, S., S. A. Byers, J. P. Price, V. T. Nguyen, O. Bensaude, D. H. Price, and W. Maury. 2007. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib, and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology 4:47. - PMC - PubMed
-
- Brenchley, J. M., B. J. Hill, D. R. Ambrozak, D. A. Price, F. J. Guenaga, J. P. Casazza, J. Kuruppu, J. Yazdani, S. A. Migueles, M. Connors, M. Roederer, D. C. Douek, and R. A. Koup. 2004. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J. Virol. 78:1160-1168. - PMC - PubMed
-
- Brooks, D. G., S. G. Kitchen, C. M. Kitchen, D. D. Scripture-Adams, and J. A. Zack. 2001. Generation of HIV latency during thymopoiesis. Nat. Med. 7:459-464. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

