Two novel families of plasmids from hyperthermophilic archaea encoding new families of replication proteins
- PMID: 20403814
- PMCID: PMC2926602
- DOI: 10.1093/nar/gkq236
Two novel families of plasmids from hyperthermophilic archaea encoding new families of replication proteins
Abstract
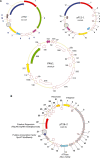
Thermococcales (phylum Euryarchaeota) are model organisms for physiological and molecular studies of hyperthermophiles. Here we describe three new plasmids from Thermococcales that could provide new tools and model systems for genetic and molecular studies in Archaea. The plasmids pTN2 from Thermococcus nautilus sp. 30-1 and pP12-1 from Pyrococcus sp. 12-1 belong to the same family. They have similar size (approximately 12 kb) and share six genes, including homologues of genes encoded by the virus PAV1 from Pyrococcus abyssi. The plasmid pT26-2 from Thermococcus sp. 26-2 (21.5 kb), that corresponds to another plasmid family, encodes many proteins having homologues in virus-like elements integrated in several genomes of Thermococcales and Methanococcales. Our analyses confirm that viruses and plasmids are evolutionary related and co-evolve with their hosts. Whereas all plasmids previously isolated from Thermococcales replicate by the rolling circle mechanism, the three plasmids described here probably replicate by the theta mechanism. The plasmids pTN2 and pP12-1 encode a putative helicase of the SFI superfamily and a new family of DNA polymerase, whose activity was demonstrated in vitro, whereas pT26-2 encodes a putative new type of helicase. This strengthens the idea that plasmids and viruses are a reservoir of novel protein families involved in DNA replication.
Figures

Similar articles
-
Insights into dynamics of mobile genetic elements in hyperthermophilic environments from five new Thermococcus plasmids.PLoS One. 2013;8(1):e49044. doi: 10.1371/journal.pone.0049044. Epub 2013 Jan 11. PLoS One. 2013. PMID: 23326305 Free PMC article.
-
Plasmids, viruses and virus-like membrane vesicles from Thermococcales.Biochem Soc Trans. 2011 Jan;39(1):36-44. doi: 10.1042/BST0390036. Biochem Soc Trans. 2011. PMID: 21265744 Review.
-
pAMT11, a novel plasmid isolated from a Thermococcus sp. strain closely related to the virus-like integrated element TKV1 of the Thermococcus kodakaraensis genome.Res Microbiol. 2011 Feb-Mar;162(2):132-43. doi: 10.1016/j.resmic.2010.11.003. Epub 2010 Dec 7. Res Microbiol. 2011. PMID: 21144896
-
A protein encoded by a new family of mobile elements from Euryarchaea exhibits three domains with novel folds.Protein Sci. 2009 Apr;18(4):850-5. doi: 10.1002/pro.73. Protein Sci. 2009. PMID: 19319959 Free PMC article.
-
Genetic elements of Thermococcales.Biochem Soc Trans. 2004 Apr;32(Pt 2):184-7. doi: 10.1042/bst0320184. Biochem Soc Trans. 2004. PMID: 15046568 Review.
Cited by
-
Genetic studies on the virus-like regions in the genome of hyperthermophilic archaeon, Thermococcus kodakarensis.Extremophiles. 2013 Jan;17(1):153-60. doi: 10.1007/s00792-012-0504-6. Epub 2012 Dec 9. Extremophiles. 2013. PMID: 23224520
-
Insights into dynamics of mobile genetic elements in hyperthermophilic environments from five new Thermococcus plasmids.PLoS One. 2013;8(1):e49044. doi: 10.1371/journal.pone.0049044. Epub 2013 Jan 11. PLoS One. 2013. PMID: 23326305 Free PMC article.
-
WASPS: web-assisted symbolic plasmid synteny server.Bioinformatics. 2020 Mar 1;36(5):1629-1631. doi: 10.1093/bioinformatics/btz745. Bioinformatics. 2020. PMID: 31589313 Free PMC article.
-
Provirus induction in hyperthermophilic archaea: characterization of Aeropyrum pernix spindle-shaped virus 1 and Aeropyrum pernix ovoid virus 1.J Bacteriol. 2011 Oct;193(19):5412-9. doi: 10.1128/JB.05101-11. Epub 2011 Jul 22. J Bacteriol. 2011. PMID: 21784945 Free PMC article.
-
Archaeal extrachromosomal genetic elements.Microbiol Mol Biol Rev. 2015 Mar;79(1):117-52. doi: 10.1128/MMBR.00042-14. Microbiol Mol Biol Rev. 2015. PMID: 25694123 Free PMC article. Review.
References
-
- Lipps G, editor. Plasmids Current Research and Future Trends. Norfolk, UK: Caaister Academic Press; 2008. p. 259.
-
- Soppa J. From genomes to function: haloarchaea as model organisms. Microbiology. 2006;152:585–590. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

