Role of SUMO:SIM-mediated protein-protein interaction in non-homologous end joining
- PMID: 20400978
- PMCID: PMC2891878
- DOI: 10.1038/onc.2010.108
Role of SUMO:SIM-mediated protein-protein interaction in non-homologous end joining
Abstract
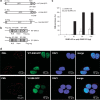
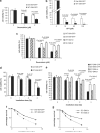
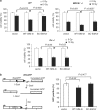
Although post-translational modifications by the small ubiquitin-like modifiers (SUMO) are known to be important in DNA damage response, it is unclear whether they have a role in double-strand break (DSB) repair by non-homologous end joining (NHEJ). Here, we analyzed various DSB repair pathways upon inhibition of SUMO-mediated protein-protein interactions using peptides that contain the SUMO-interaction motif (SIM) and discriminate between mono- and SUMO-chain modifications. The SIM peptides specifically inhibit NHEJ as shown by in vivo repair assays and radio-sensitivity of cell lines deficient in different DSB repair pathways. Furthermore, mono-SUMO, instead of SUMO-chain, modifications appear to be involved in NHEJ. Immunoprecipitation experiments also showed that the SIM peptide interacted with SUMOylated Ku70 after radiation. This study is the first to show an important role for SUMO:SIM-mediated protein-protein interactions in NHEJ, and provides a mechanistic basis for the role of SIM peptide in sensitizing genotoxic stress of cancer cells.
Figures


Similar articles
-
Global non-covalent SUMO interaction networks reveal SUMO-dependent stabilization of the non-homologous end joining complex.Cell Rep. 2021 Jan 26;34(4):108691. doi: 10.1016/j.celrep.2021.108691. Cell Rep. 2021. PMID: 33503430
-
Small ubiquitin-like modifier (SUMO) isoforms and conjugation-independent function in DNA double-strand break repair pathways.J Biol Chem. 2014 Aug 1;289(31):21289-95. doi: 10.1074/jbc.C114.582122. Epub 2014 Jun 25. J Biol Chem. 2014. PMID: 24966330 Free PMC article.
-
SUMO Interacting Motifs: Structure and Function.Cells. 2021 Oct 21;10(11):2825. doi: 10.3390/cells10112825. Cells. 2021. PMID: 34831049 Free PMC article. Review.
-
Pro-recombination Role of Srs2 Protein Requires SUMO (Small Ubiquitin-like Modifier) but Is Independent of PCNA (Proliferating Cell Nuclear Antigen) Interaction.J Biol Chem. 2016 Apr 1;291(14):7594-607. doi: 10.1074/jbc.M115.685891. Epub 2016 Feb 9. J Biol Chem. 2016. PMID: 26861880 Free PMC article.
-
Targeting DNA Double-Strand Break (DSB) Repair to Counteract Tumor Radio-resistance.Curr Drug Targets. 2019;20(9):891-902. doi: 10.2174/1389450120666190222181857. Curr Drug Targets. 2019. PMID: 30806313 Review.
Cited by
-
Biochemical analysis of protein SUMOylation.Curr Protoc Mol Biol. 2012 Jul;Chapter 10:Unit10.29. doi: 10.1002/0471142727.mb1029s99. Curr Protoc Mol Biol. 2012. PMID: 22870855 Free PMC article.
-
Design of high-throughput screening assays and identification of a SUMO1-specific small molecule chemotype targeting the SUMO-interacting motif-binding surface.ACS Comb Sci. 2015 Apr 13;17(4):239-46. doi: 10.1021/co500181b. Epub 2015 Mar 23. ACS Comb Sci. 2015. PMID: 25719760 Free PMC article.
-
Adhiron: a stable and versatile peptide display scaffold for molecular recognition applications.Protein Eng Des Sel. 2014 May;27(5):145-55. doi: 10.1093/protein/gzu007. Epub 2014 Mar 25. Protein Eng Des Sel. 2014. PMID: 24668773 Free PMC article.
-
Oncogene UBE2I enhances cellular invasion, migration and proliferation abilities via autophagy-related pathway resulting in poor prognosis in hepatocellular carcinoma.Am J Cancer Res. 2020 Dec 1;10(12):4178-4197. eCollection 2020. Am J Cancer Res. 2020. PMID: 33414994 Free PMC article.
-
Microarray screening reveals two non-conventional SUMO-binding modules linked to DNA repair by non-homologous end-joining.Nucleic Acids Res. 2022 May 6;50(8):4732-4754. doi: 10.1093/nar/gkac237. Nucleic Acids Res. 2022. PMID: 35420136 Free PMC article.
References
-
- Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. - PubMed
-
- Carson JP, Zhang N, Frampton GM, Gerry NP, Lenburg ME, Christman MF. Pharmacogenomic identification of targets for adjuvant therapy with the topoisomerase poison camptothecin. Cancer Res. 2004;64:2096–2104. - PubMed
-
- Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–2319. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32CA134180/CA/NCI NIH HHS/United States
- R01 GM086171/GM/NIGMS NIH HHS/United States
- R01 CA120954/CA/NCI NIH HHS/United States
- F32 CA134180-03/CA/NCI NIH HHS/United States
- R01 GM074748-03/GM/NIGMS NIH HHS/United States
- F32 CA134180-02/CA/NCI NIH HHS/United States
- F32 CA134180/CA/NCI NIH HHS/United States
- R37 CA050519/CA/NCI NIH HHS/United States
- R01 GM074748-04/GM/NIGMS NIH HHS/United States
- R01CA120954/CA/NCI NIH HHS/United States
- R01 GM086171-04/GM/NIGMS NIH HHS/United States
- R01GM086171/GM/NIGMS NIH HHS/United States
- R01GM074748/GM/NIGMS NIH HHS/United States
- R37CA050519-20/CA/NCI NIH HHS/United States
- R01 CA120954-04/CA/NCI NIH HHS/United States
- R01 DE014183/DE/NIDCR NIH HHS/United States
- R01DE14183/DE/NIDCR NIH HHS/United States
- R01 GM086171-03/GM/NIGMS NIH HHS/United States
- F32 CA134180-01/CA/NCI NIH HHS/United States
- R01 GM074748/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

