LKB1 suppresses p21-activated kinase-1 (PAK1) by phosphorylation of Thr109 in the p21-binding domain
- PMID: 20400510
- PMCID: PMC2881753
- DOI: 10.1074/jbc.M109.079137
LKB1 suppresses p21-activated kinase-1 (PAK1) by phosphorylation of Thr109 in the p21-binding domain
Abstract
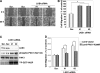
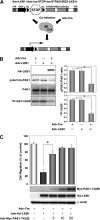
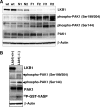
The serine/threonine protein kinase LKB1 is a tumor suppressor gene mutated in Peutz-Jeghers syndrome patients. The mutations are found also in several types of sporadic cancer. Although LKB1 is implicated in suppression of cell growth and metastasis, the detailed mechanisms have not yet been elucidated. In this study, we investigated the effect of LKB1 on cell motility, whose acquisition occurs in early metastasis. The knockdown of LKB1 enhanced cell migration and PAK1 activity in human colon cancer HCT116 cells, whereas forced expression of LKB1 in Lkb1-null mouse embryonic fibroblasts suppressed PAK1 activity and PAK1-mediated cell migration simultaneously. Notably, LKB1 directly phosphorylated PAK1 at Thr(109) in the p21-binding domain in vitro. The phosphomimetic T109E mutant showed significantly lower protein kinase activity than wild-type PAK1, suggesting that the phosphorylation at Thr(109) by LKB1 was responsible for suppression of PAK1. Consistently, the nonphosphorylatable T109A mutant was resistant to suppression by LKB1. Furthermore, we found that PAK1 was activated in the hepatocellular carcinomas and the precancerous liver lesions of Lkb1(+/-) mice. Taken together, these results suggest that PAK1 is a direct downstream target of LKB1 and plays an essential role in LKB1-induced suppression of cell migration.
Figures



Similar articles
-
STE20-related kinase adaptor protein α (STRADα) regulates cell polarity and invasion through PAK1 signaling in LKB1-null cells.J Biol Chem. 2012 May 25;287(22):18758-68. doi: 10.1074/jbc.M111.316422. Epub 2012 Apr 6. J Biol Chem. 2012. PMID: 22493453 Free PMC article.
-
Synapses of amphids defective (SAD-A) kinase promotes glucose-stimulated insulin secretion through activation of p21-activated kinase (PAK1) in pancreatic β-Cells.J Biol Chem. 2012 Jul 27;287(31):26435-44. doi: 10.1074/jbc.M112.378372. Epub 2012 Jun 5. J Biol Chem. 2012. PMID: 22669945 Free PMC article.
-
P21-activated protein kinase is overexpressed in hepatocellular carcinoma and enhances cancer metastasis involving c-Jun NH2-terminal kinase activation and paxillin phosphorylation.Cancer Res. 2007 Apr 15;67(8):3601-8. doi: 10.1158/0008-5472.CAN-06-3994. Cancer Res. 2007. PMID: 17440071
-
Tyrosyl phosphorylated serine-threonine kinase PAK1 is a novel regulator of prolactin-dependent breast cancer cell motility and invasion.Adv Exp Med Biol. 2015;846:97-137. doi: 10.1007/978-3-319-12114-7_5. Adv Exp Med Biol. 2015. PMID: 25472536 Free PMC article. Review.
-
Molecular mechanisms of tumor suppression by LKB1.FEBS Lett. 2011 Apr 6;585(7):944-51. doi: 10.1016/j.febslet.2010.12.034. Epub 2010 Dec 27. FEBS Lett. 2011. PMID: 21192934 Review.
Cited by
-
Altered LKB1/AMPK/TSC1/TSC2/mTOR signaling causes disruption of Sertoli cell polarity and spermatogenesis.Hum Mol Genet. 2012 Oct 15;21(20):4394-405. doi: 10.1093/hmg/dds272. Epub 2012 Jul 12. Hum Mol Genet. 2012. PMID: 22791749 Free PMC article.
-
Republished: tracing PAKs from GI inflammation to cancer.Postgrad Med J. 2014 Nov;90(1069):657-68. doi: 10.1136/postgradmedj-2014-306768rep. Postgrad Med J. 2014. PMID: 25335797 Free PMC article.
-
LKB1 signaling in advancing cell differentiation.Fam Cancer. 2011 Sep;10(3):425-35. doi: 10.1007/s10689-011-9441-2. Fam Cancer. 2011. PMID: 21519908 Review.
-
Liver kinase B1 (LKB1) in the pathogenesis of epithelial cancers.Cancer Lett. 2011 Jul 1;306(1):1-9. doi: 10.1016/j.canlet.2011.01.014. Epub 2011 Mar 29. Cancer Lett. 2011. PMID: 21450399 Free PMC article. Review.
-
Liver Kinase B1 Functions as a Regulator for Neural Development and a Therapeutic Target for Neural Repair.Cells. 2022 Sep 14;11(18):2861. doi: 10.3390/cells11182861. Cells. 2022. PMID: 36139438 Free PMC article. Review.
References
-
- Giardiello F. M., Welsh S. B., Hamilton S. R., Offerhaus G. J., Gittelsohn A. M., Booker S. V., Krush A. J., Yardley J. H., Luk G. D. (1987) N. Engl. J. Med. 316, 1511–1514 - PubMed
-
- Hemminki A., Markie D., Tomlinson I., Avizienyte E., Roth S., Loukola A., Bignell G., Warren W., Aminoff M., Höglund P., Järvinen H., Kristo P., Pelin K., Ridanpää M., Salovaara R., Toro T., Bodmer W., Olschwang S., Olsen A. S., Stratton M. R., de la Chapelle A., Aaltonen L. A. (1998) Nature 391, 184–187 - PubMed
-
- Jenne D. E., Reimann H., Nezu J., Friedel W., Loff S., Jeschke R., Müller O., Back W., Zimmer M. (1998) Nat. Genet. 18, 38–43 - PubMed
-
- Launonen V. (2005) Hum. Mutat. 26, 291–297 - PubMed
-
- Alessi D. R., Sakamoto K., Bayascas J. R. (2006) Annu. Rev. Biochem. 75, 137–163 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

