Regulation of microtubule dynamics by Bim1 and Bik1, the budding yeast members of the EB1 and CLIP-170 families of plus-end tracking proteins
- PMID: 20392838
- PMCID: PMC2883945
- DOI: 10.1091/mbc.e10-02-0083
Regulation of microtubule dynamics by Bim1 and Bik1, the budding yeast members of the EB1 and CLIP-170 families of plus-end tracking proteins
Abstract
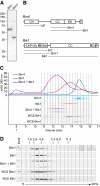
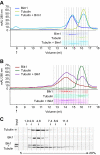
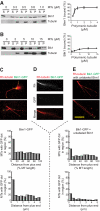
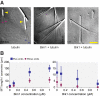
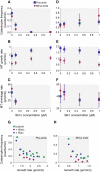
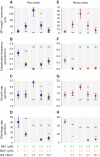
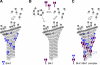
Microtubule dynamics are regulated by plus-end tracking proteins (+TIPs), which bind microtubule ends and influence their polymerization properties. In addition to binding microtubules, most +TIPs physically associate with other +TIPs, creating a complex web of interactions. To fully understand how +TIPs regulate microtubule dynamics, it is essential to know the intrinsic biochemical activities of each +TIP and how +TIP interactions affect these activities. Here, we describe the activities of Bim1 and Bik1, two +TIP proteins from budding yeast and members of the EB1 and CLIP-170 families, respectively. We find that purified Bim1 and Bik1 form homodimers that interact with each other to form a tetramer. Bim1 binds along the microtubule lattice but with highest affinity for the microtubule end; however, Bik1 requires Bim1 for localization to the microtubule lattice and end. In vitro microtubule polymerization assays show that Bim1 promotes microtubule assembly, primarily by decreasing the frequency of catastrophes. In contrast, Bik1 inhibits microtubule assembly by slowing growth and, consequently, promoting catastrophes. Interestingly, the Bim1-Bik1 complex affects microtubule dynamics in much the same way as Bim1 alone. These studies reveal new activities for EB1 and CLIP-170 family members and demonstrate how interactions between two +TIP proteins influence their activities.
Figures
Similar articles
-
Structure-Function Relationship of the Bik1-Bim1 Complex.Structure. 2018 Apr 3;26(4):607-618.e4. doi: 10.1016/j.str.2018.03.003. Epub 2018 Mar 22. Structure. 2018. PMID: 29576319
-
The regulation of microtubule dynamics in Saccharomyces cerevisiae by three interacting plus-end tracking proteins.Mol Biol Cell. 2006 Jun;17(6):2789-98. doi: 10.1091/mbc.e05-09-0892. Epub 2006 Mar 29. Mol Biol Cell. 2006. PMID: 16571681 Free PMC article.
-
Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1.Mol Cell. 2007 Sep 21;27(6):976-91. doi: 10.1016/j.molcel.2007.07.023. Mol Cell. 2007. PMID: 17889670 Free PMC article.
-
Structural and mechanistic insights into microtubule end-binding proteins.Curr Opin Cell Biol. 2010 Feb;22(1):88-95. doi: 10.1016/j.ceb.2009.10.009. Epub 2009 Dec 1. Curr Opin Cell Biol. 2010. PMID: 19959349 Review.
-
CLIP-170 family members: a motor-driven ride to microtubule plus ends.Dev Cell. 2004 Jun;6(6):746-8. doi: 10.1016/j.devcel.2004.05.017. Dev Cell. 2004. PMID: 15177023 Review.
Cited by
-
Mitotic spindle form and function.Genetics. 2012 Apr;190(4):1197-224. doi: 10.1534/genetics.111.128710. Genetics. 2012. PMID: 22491889 Free PMC article.
-
Spatial control of microtubule length and lifetime by opposing stabilizing and destabilizing functions of Kinesin-8.Curr Biol. 2014 Aug 18;24(16):1826-35. doi: 10.1016/j.cub.2014.06.069. Epub 2014 Jul 31. Curr Biol. 2014. PMID: 25088560 Free PMC article.
-
In vivo roles of the basic domain of dynactin p150 in microtubule plus-end tracking and dynein function.Traffic. 2012 Mar;13(3):375-87. doi: 10.1111/j.1600-0854.2011.01312.x. Epub 2011 Dec 18. Traffic. 2012. PMID: 22106867 Free PMC article.
-
Emerging roles of sumoylation in the regulation of actin, microtubules, intermediate filaments, and septins.Cytoskeleton (Hoboken). 2015 Jul;72(7):305-39. doi: 10.1002/cm.21226. Epub 2015 Aug 22. Cytoskeleton (Hoboken). 2015. PMID: 26033929 Free PMC article. Review.
-
Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels.Nat Cell Biol. 2013 Jun;15(6):688-93. doi: 10.1038/ncb2744. Epub 2013 May 12. Nat Cell Biol. 2013. PMID: 23666085
References
-
- Akhmanova A., Hoogenraad C. C. Microtubule plus-end-tracking proteins: mechanisms and functions. Curr. Opin. Cell Biol. 2005;1:47–54. - PubMed
-
- Akhmanova A., Steinmetz M. O. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 2008;4:309–322. - PubMed
-
- Arnal I., Heichette C., Diamantopoulos G. S., Chretien D. CLIP-170/tubulin-curved oligomers coassemble at microtubule ends and promote rescues. Curr. Biol. 2004;23:2086–2095. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

