NUDT16 is a (deoxy)inosine diphosphatase, and its deficiency induces accumulation of single-strand breaks in nuclear DNA and growth arrest
- PMID: 20385596
- PMCID: PMC2919730
- DOI: 10.1093/nar/gkq249
NUDT16 is a (deoxy)inosine diphosphatase, and its deficiency induces accumulation of single-strand breaks in nuclear DNA and growth arrest
Abstract
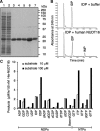
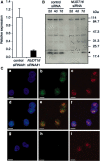
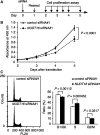
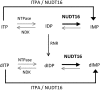
Nucleotides function in a variety of biological reactions; however, they can undergo various chemical modifications. Such modified nucleotides may be toxic to cells if not eliminated from the nucleotide pools. We performed a screen for modified-nucleotide binding proteins and identified human nucleoside diphosphate linked moiety X-type motif 16 (NUDT16) protein as an inosine triphosphate (ITP)/xanthosine triphosphate (XTP)/GTP-binding protein. Recombinant NUDT16 hydrolyzes purine nucleoside diphosphates to the corresponding nucleoside monophosphates. Among 29 nucleotides examined, the highest k(cat)/K(m) values were for inosine diphosphate (IDP) and deoxyinosine diphosphate (dIDP). Moreover, NUDT16 moderately hydrolyzes (deoxy)inosine triphosphate ([d]ITP). NUDT16 is mostly localized in the nucleus, and especially in the nucleolus. Knockdown of NUDT16 in HeLa MR cells caused cell cycle arrest in S-phase, reduced cell proliferation, increased accumulation of single-strand breaks in nuclear DNA as well as increased levels of inosine in RNA. We thus concluded that NUDT16 is a (deoxy)inosine diphosphatase that may function mainly in the nucleus to protect cells from deleterious effects of (d)ITP.
Figures


Similar articles
-
NUDT16 and ITPA play a dual protective role in maintaining chromosome stability and cell growth by eliminating dIDP/IDP and dITP/ITP from nucleotide pools in mammals.Nucleic Acids Res. 2010 May;38(9):2891-903. doi: 10.1093/nar/gkp1250. Epub 2010 Jan 15. Nucleic Acids Res. 2010. PMID: 20081199 Free PMC article.
-
A comprehensive screening system for damaged nucleotide-binding proteins.Mutat Res. 2010 Nov 28;703(1):37-42. doi: 10.1016/j.mrgentox.2010.06.005. Epub 2010 Jun 11. Mutat Res. 2010. PMID: 20542141 Review.
-
ITPA (inosine triphosphate pyrophosphatase): from surveillance of nucleotide pools to human disease and pharmacogenetics.Mutat Res. 2013 Oct-Dec;753(2):131-146. doi: 10.1016/j.mrrev.2013.08.001. Epub 2013 Aug 19. Mutat Res. 2013. PMID: 23969025 Free PMC article. Review.
-
Inosine Triphosphate Pyrophosphatase (ITPase): Functions, Mutations, Polymorphisms and Its Impact on Cancer Therapies.Cells. 2022 Jan 24;11(3):384. doi: 10.3390/cells11030384. Cells. 2022. PMID: 35159194 Free PMC article. Review.
-
Cloning, expression, and characterization of a human inosine triphosphate pyrophosphatase encoded by the itpa gene.J Biol Chem. 2001 Jun 1;276(22):18695-701. doi: 10.1074/jbc.M011084200. Epub 2001 Mar 13. J Biol Chem. 2001. PMID: 11278832
Cited by
-
8-Oxoguanine causes neurodegeneration during MUTYH-mediated DNA base excision repair.J Clin Invest. 2012 Dec;122(12):4344-61. doi: 10.1172/JCI65053. Epub 2012 Nov 12. J Clin Invest. 2012. PMID: 23143307 Free PMC article.
-
Potent and Selective Inhibitors of 8-Oxoguanine DNA Glycosylase.J Am Chem Soc. 2018 Feb 14;140(6):2105-2114. doi: 10.1021/jacs.7b09316. Epub 2018 Feb 5. J Am Chem Soc. 2018. PMID: 29376367 Free PMC article.
-
Structural Basis for the Specificity of Human NUDT16 and Its Regulation by Inosine Monophosphate.PLoS One. 2015 Jun 29;10(6):e0131507. doi: 10.1371/journal.pone.0131507. eCollection 2015. PLoS One. 2015. PMID: 26121039 Free PMC article.
-
Hydrolytic activity of human Nudt16 enzyme on dinucleotide cap analogs and short capped oligonucleotides.RNA. 2018 May;24(5):633-642. doi: 10.1261/rna.065698.118. Epub 2018 Feb 26. RNA. 2018. PMID: 29483298 Free PMC article.
-
Multiple Nudix family proteins possess mRNA decapping activity.RNA. 2013 Mar;19(3):390-9. doi: 10.1261/rna.037309.112. Epub 2013 Jan 25. RNA. 2013. PMID: 23353937 Free PMC article.
References
-
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair And Mutagenesis. 2nd edn. Washington: ASM Press; 2005.
-
- Nakabeppu Y, Sakumi K, Sakamoto K, Tsuchimoto D, Tsuzuki T, Nakatsu Y. Mutagenesis and carcinogenesis caused by the oxidation of nucleic acids. Biol. Chem. 2006;387:373–379. - PubMed
-
- Nakabeppu Y, Kajitani K, Sakamoto K, Yamaguchi H, Tsuchimoto D. MTH1, an oxidized purine nucleoside triphosphatase, prevents the cytotoxicity and neurotoxicity of oxidized purine nucleotides. DNA Repair. 2006;5:761–772. - PubMed
-
- Nakabeppu Y, Tsuchimoto D, Yamaguchi H, Sakumi K. Oxidative damage in nucleic acids and Parkinson’s disease. J. Neurosci. Res. 2007;85:919–934. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

