Direct regulation of Bacillus subtilis phoPR transcription by transition state regulator ScoC
- PMID: 20382764
- PMCID: PMC2901694
- DOI: 10.1128/JB.00089-10
Direct regulation of Bacillus subtilis phoPR transcription by transition state regulator ScoC
Abstract
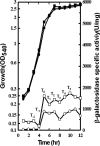
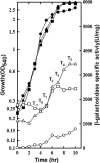
Induction of the Pho response in Bacillus subtilis occurs when the P(i) concentrations in the growth medium fall below 0.1 mM, a condition which results in slowed cellular growth followed by entry into stationary phase. The phoPR promoter region contains three sigma(A)-responsive promoters; only promoter P(A4) is PhoP autoregulated. Expression of the phoPR operon is postexponential, suggesting the possibility of a repressor role for a transition-state-regulatory protein(s). Expression of a phoPR promoter-lacZ fusion in a scoC loss-of-function mutant strain grown in low-phosphate defined medium was significantly higher than expression in the wild-type strain during exponential growth or stationary phase. Derepression in the scoC strain from a phoP promoter fusion containing a mutation in the CcpA binding site (cre1) was further elevated approximately 1.4-fold, indicating that the repressor effects of ScoC and CcpA on phoP expression were cumulative. DNase I footprinting showed protection of putative binding sites by ScoC, which included the -10 and/or -35 elements of five (P(B1), P(E2), P(A3), P(A4), and P(A6)) of the six promoters within the phoPR promoter region. P(A6) was expressed in vivo from the phoP cre1 promoter fusion in both wild-type and scoC strains. Evidence for ScoC repression in vivo was shown by primer extension for P(A4) and P(A3) from the wild-type promoter and for P(A4) and P(E2) from the phoP cre1 promoter. The latter may reflect ScoC repression of sporulation that indirectly affects phoPR transcription. ScoC was shown to repress P(A6), P(A4), P(E2), and P(B1) in vitro.
Figures

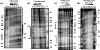


Similar articles
-
CcpA causes repression of the phoPR promoter through a novel transcription start site, P(A6).J Bacteriol. 2006 Feb;188(4):1266-78. doi: 10.1128/JB.188.4.1266-1278.2006. J Bacteriol. 2006. PMID: 16452408 Free PMC article.
-
Bacillus subtilis phosphorylated PhoP: direct activation of the E(sigma)A- and repression of the E(sigma)E-responsive phoB-PS+V promoters during pho response.J Bacteriol. 2005 Aug;187(15):5166-78. doi: 10.1128/JB.187.15.5166-5178.2005. J Bacteriol. 2005. PMID: 16030210 Free PMC article.
-
Autoinduction of Bacillus subtilis phoPR operon transcription results from enhanced transcription from EsigmaA- and EsigmaE-responsive promoters by phosphorylated PhoP.J Bacteriol. 2004 Jul;186(13):4262-75. doi: 10.1128/JB.186.13.4262-4275.2004. J Bacteriol. 2004. PMID: 15205429 Free PMC article.
-
Transcriptional regulation of the phoPR operon in Bacillus subtilis.J Bacteriol. 2004 Feb;186(4):1182-90. doi: 10.1128/JB.186.4.1182-1190.2004. J Bacteriol. 2004. PMID: 14762014 Free PMC article.
-
Sequential action of two-component genetic switches regulates the PHO regulon in Bacillus subtilis.J Bacteriol. 1994 Mar;176(5):1348-58. doi: 10.1128/jb.176.5.1348-1358.1994. J Bacteriol. 1994. PMID: 8113174 Free PMC article.
Cited by
-
Phosphate (Pi) Transporter PIT1 Induces Pi Starvation in Salmonella-Containing Vacuole in HeLa Cells.Int J Mol Sci. 2023 Dec 7;24(24):17216. doi: 10.3390/ijms242417216. Int J Mol Sci. 2023. PMID: 38139044 Free PMC article.
-
Two Distinct Regulatory Systems Control Pulcherrimin Biosynthesis in Bacillus subtilis.bioRxiv [Preprint]. 2024 Jan 4:2024.01.03.574033. doi: 10.1101/2024.01.03.574033. bioRxiv. 2024. Update in: PLoS Genet. 2024 May 16;20(5):e1011283. doi: 10.1371/journal.pgen.1011283 PMID: 38260623 Free PMC article. Updated. Preprint.
-
Identification of L-Valine-initiated-germination-active genes in Bacillus subtilis using Tn-seq.PLoS One. 2019 Jun 14;14(6):e0218220. doi: 10.1371/journal.pone.0218220. eCollection 2019. PLoS One. 2019. PMID: 31199835 Free PMC article.
-
Two distinct regulatory systems control pulcherrimin biosynthesis in Bacillus subtilis.PLoS Genet. 2024 May 16;20(5):e1011283. doi: 10.1371/journal.pgen.1011283. eCollection 2024 May. PLoS Genet. 2024. PMID: 38753885 Free PMC article.
-
Analysis of a bac operon-silenced strain suggests pleiotropic effects of bacilysin in Bacillus subtilis.J Microbiol. 2020 Apr;58(4):297-313. doi: 10.1007/s12275-020-9064-0. Epub 2020 Jan 28. J Microbiol. 2020. PMID: 31989543
References
-
- Abdel-Fattah, W. R. 2007. Bacillus subtilis Pho∼P direct roles in Pho and Res regulation in response to Pi-stress. Ph.D. thesis. University of Illinois at Chicago, Chicago, IL.
-
- Balassa, G., B. Dod, V. Jeannoda, P. Milhaud, J. Zucca, J. C. Sousa, and M. T. Silva. 1978. Pleiotropic control mutations affecting the sporulation of Bacillus subtilis. Ann. Microbiol. (Paris) 129 B:537-549. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

