ADAR editing in double-stranded UTRs and other noncoding RNA sequences
- PMID: 20382028
- PMCID: PMC2897959
- DOI: 10.1016/j.tibs.2010.02.008
ADAR editing in double-stranded UTRs and other noncoding RNA sequences
Abstract
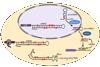
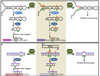
ADARs are a family of enzymes, present in all animals, that convert adenosine to inosine within double-stranded RNA (dsRNA). Inosine and adenosine have different base-pairing properties, and thus, editing alters RNA structure, coding potential and splicing patterns. The first ADAR substrates identified were edited in codons, and ADARs were presumed to function primarily in proteome diversification. Although this is an important function of ADARs, especially in the nervous system, editing in coding sequences is rare compared to editing in noncoding sequences. Introns and untranslated regions of mRNA are the primary noncoding targets, but editing also occurs in small RNAs, such as miRNAs. Although the role of editing in noncoding sequences remains unclear, ongoing research suggests functions in the regulation of a variety of post-transcriptional processes.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures
Similar articles
-
A-to-I editing of protein coding and noncoding RNAs.Crit Rev Biochem Mol Biol. 2012 Nov-Dec;47(6):493-501. doi: 10.3109/10409238.2012.714350. Epub 2012 Sep 18. Crit Rev Biochem Mol Biol. 2012. PMID: 22988838 Review.
-
Noncoding regions of C. elegans mRNA undergo selective adenosine to inosine deamination and contain a small number of editing sites per transcript.RNA Biol. 2015;12(2):162-74. doi: 10.1080/15476286.2015.1017220. RNA Biol. 2015. PMID: 25826568 Free PMC article.
-
A-to-I RNA editing - thinking beyond the single nucleotide.RNA Biol. 2017 Dec 2;14(12):1690-1694. doi: 10.1080/15476286.2017.1364830. Epub 2017 Oct 11. RNA Biol. 2017. PMID: 28820319 Free PMC article. Review.
-
ADAR-mediated RNA editing in non-coding RNA sequences.Sci China Life Sci. 2013 Oct;56(10):944-52. doi: 10.1007/s11427-013-4546-5. Epub 2013 Sep 5. Sci China Life Sci. 2013. PMID: 24008387 Review.
-
RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA.Proc Natl Acad Sci U S A. 2002 Jun 11;99(12):7906-11. doi: 10.1073/pnas.112704299. Epub 2002 Jun 4. Proc Natl Acad Sci U S A. 2002. PMID: 12048240 Free PMC article.
Cited by
-
STAU1 binding 3' UTR IRAlus complements nuclear retention to protect cells from PKR-mediated translational shutdown.Genes Dev. 2013 Jul 1;27(13):1495-510. doi: 10.1101/gad.220962.113. Genes Dev. 2013. PMID: 23824540 Free PMC article.
-
Inverted Alu dsRNA structures do not affect localization but can alter translation efficiency of human mRNAs independent of RNA editing.Nucleic Acids Res. 2012 Sep 1;40(17):8637-45. doi: 10.1093/nar/gks590. Epub 2012 Jun 25. Nucleic Acids Res. 2012. PMID: 22735697 Free PMC article.
-
Protein-Encoding RNA to RNA Information Transfer in Mammalian Cells: RNA-dependent mRNA Amplification. Identification of Chimeric RNA Intermediates and Putative RNA End Products.Ann Integr Mol Med. 2019;1(1):23-47. Epub 2019 Aug 22. Ann Integr Mol Med. 2019. PMID: 31656957 Free PMC article.
-
The dsRBP and inactive editor ADR-1 utilizes dsRNA binding to regulate A-to-I RNA editing across the C. elegans transcriptome.Cell Rep. 2014 Feb 27;6(4):599-607. doi: 10.1016/j.celrep.2014.01.011. Epub 2014 Feb 6. Cell Rep. 2014. PMID: 24508457 Free PMC article.
-
Structure-mediated modulation of mRNA abundance by A-to-I editing.Nat Commun. 2017 Nov 2;8(1):1255. doi: 10.1038/s41467-017-01459-7. Nat Commun. 2017. PMID: 29093448 Free PMC article.
References
-
- Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 2000;34:499–531. - PubMed
-
- Maydanovych O, Beal PA. Breaking the central dogma by RNA editing. Chem. Rev. 2006;106:3397–3411. - PubMed
-
- Wang Q, et al. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

