Role of vimA in cell surface biogenesis in Porphyromonas gingivalis
- PMID: 20378652
- PMCID: PMC3068682
- DOI: 10.1099/mic.0.038331-0
Role of vimA in cell surface biogenesis in Porphyromonas gingivalis
Abstract
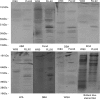
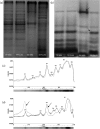
The Porphyromonas gingivalis vimA gene has been previously shown to play a significant role in the biogenesis of gingipains. Further, in P. gingivalis FLL92, a vimA-defective mutant, there was increased auto-aggregation, suggesting alteration in membrane surface proteins. In order to determine the role of the VimA protein in cell surface biogenesis, the surface morphology of P. gingivalis FLL92 was further characterized. Transmission electron microscopy demonstrated abundant fimbrial appendages and a less well defined and irregular capsule in FLL92 compared with the wild-type. In addition, atomic force microscopy showed that the wild-type had a smoother surface compared with FLL92. Western blot analysis using anti-FimA antibodies showed a 41 kDa immunoreactive protein band in P. gingivalis FLL92 which was missing in the wild-type P. gingivalis W83 strain. There was increased sensitivity to globomycin and vancomycin in FLL92 compared with the wild-type. Outer membrane fractions from FLL92 had a modified lectin-binding profile. Furthermore, in contrast with the wild-type strain, nine proteins were missing from the outer membrane fraction of FLL92, while 20 proteins present in that fraction from FLL92 were missing in the wild-type strain. Taken together, these results suggest that the VimA protein affects capsular synthesis and fimbrial phenotypic expression, and plays a role in the glycosylation and anchorage of several surface proteins.
Figures
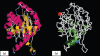


Comment in
-
Controlling Porphyromonas gingivalis requires Vim.Microbiology (Reading). 2010 Jul;156(Pt 7):1907-1908. doi: 10.1099/mic.0.041251-0. Epub 2010 May 13. Microbiology (Reading). 2010. PMID: 20466766 No abstract available.
Similar articles
-
Altered gingipain maturation in vimA- and vimE-defective isogenic mutants of Porphyromonas gingivalis.Infect Immun. 2005 Mar;73(3):1357-66. doi: 10.1128/IAI.73.3.1357-1366.2005. Infect Immun. 2005. PMID: 15731033 Free PMC article.
-
Role of Acetyltransferase PG1842 in Gingipain Biogenesis in Porphyromonas gingivalis.J Bacteriol. 2018 Nov 26;200(24):e00385-18. doi: 10.1128/JB.00385-18. Print 2018 Dec 15. J Bacteriol. 2018. PMID: 30249709 Free PMC article.
-
Gingipain RgpB is excreted as a proenzyme in the vimA-defective mutant Porphyromonas gingivalis FLL92.Infect Immun. 2003 Jul;71(7):3740-7. doi: 10.1128/IAI.71.7.3740-3747.2003. Infect Immun. 2003. PMID: 12819055 Free PMC article.
-
VimA mediates multiple functions that control virulence in Porphyromonas gingivalis.Mol Oral Microbiol. 2013 Jun;28(3):167-80. doi: 10.1111/omi.12017. Epub 2012 Dec 21. Mol Oral Microbiol. 2013. PMID: 23279905 Free PMC article. Review.
-
Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen.FEMS Microbiol Lett. 2012 Aug;333(1):1-9. doi: 10.1111/j.1574-6968.2012.02579.x. Epub 2012 May 28. FEMS Microbiol Lett. 2012. PMID: 22530835 Review.
Cited by
-
Metabolome variations in the Porphyromonas gingivalis vimA mutant during hydrogen peroxide-induced oxidative stress.Mol Oral Microbiol. 2015 Apr;30(2):111-27. doi: 10.1111/omi.12075. Epub 2014 Oct 16. Mol Oral Microbiol. 2015. PMID: 25055986 Free PMC article.
-
Oxidative stress resistance in Porphyromonas gingivalis.Future Microbiol. 2012 Apr;7(4):497-512. doi: 10.2217/fmb.12.17. Future Microbiol. 2012. PMID: 22439726 Free PMC article. Review.
-
VimA-dependent modulation of the secretome in Porphyromonas gingivalis.Mol Oral Microbiol. 2012 Dec;27(6):420-35. doi: 10.1111/j.2041-1014.2012.00661.x. Epub 2012 Aug 9. Mol Oral Microbiol. 2012. PMID: 23134608 Free PMC article.
-
Quercetin inhibits virulence properties of Porphyromas gingivalis in periodontal disease.Sci Rep. 2020 Oct 27;10(1):18313. doi: 10.1038/s41598-020-74977-y. Sci Rep. 2020. PMID: 33110205 Free PMC article.
-
Sialidase and sialoglycoproteases can modulate virulence in Porphyromonas gingivalis.Infect Immun. 2011 Jul;79(7):2779-91. doi: 10.1128/IAI.00106-11. Epub 2011 Apr 18. Infect Immun. 2011. PMID: 21502589 Free PMC article.
References
-
- Amano, A., Kuboniwa, M., Nakagawa, I., Akiyama, S., Morisaki, I. & Hamada, S. (2000). Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J Dent Res 79, 1664–1668. - PubMed
-
- Amano, A., Nakagawa, I., Okahashi, N. & Hamada, N. (2004). Variations of Porphyromonas gingivalis fimbriae in relation to microbial pathogenesis. J Periodontal Res 39, 136–142. - PubMed
-
- Athavankar, S. & Peterson, B. R. (2003). Control of gene expression with small molecules: biotin-mediated acylation of targeted lysine residues in recombinant yeast. Chem Biol 10, 1245–1253. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

