Multiple molecules of Hsc70 and a dimer of DjA1 independently bind to an unfolded protein
- PMID: 20363747
- PMCID: PMC2878007
- DOI: 10.1074/jbc.M110.101501
Multiple molecules of Hsc70 and a dimer of DjA1 independently bind to an unfolded protein
Abstract
Protein folding is a prominent chaperone function of the Hsp70 system. Refolding of an unfolded protein is efficiently mediated by the Hsc70 system with either type 1 DnaJ protein, DjA1 or DjA2, and a nucleotide exchange factor. A surface plasmon resonance technique was applied to investigate substrate recognition by the Hsc70 system and demonstrated that multiple Hsc70 proteins and a dimer of DjA1 initially bind independently to an unfolded protein. The association rate of the Hsc70 was faster than that of DjA1 under folding-compatible conditions. The Hsc70 binding involved a conformational change, whereas the DjA1 binding was bivalent and substoichiometric. Consistently, we found that the bound (14)C-labeled Hsc70 to the unfolded protein became more resistant to tryptic digestion. The gel filtration and cross-linking experiments revealed the predominant presence of the DjA1 dimer. Furthermore, the Hsc70 and DjA1 bound to distinct sets of peptide array sequences. All of these findings argue against the generality of the widely proposed hypothesis that the DnaJ-bound substrate is targeted and transferred to Hsp70. Instead, these results suggest the importance of the bivalent binding of DjA1 dimer that limits unfavorable transitions of substrate conformations in protein folding.
Figures
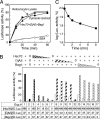




Similar articles
-
The DNAJA2 substrate release mechanism is essential for chaperone-mediated folding.J Biol Chem. 2012 Dec 7;287(50):41939-54. doi: 10.1074/jbc.M112.413278. Epub 2012 Oct 22. J Biol Chem. 2012. PMID: 23091061 Free PMC article.
-
Functional divergence between co-chaperones of Hsc70.J Biol Chem. 2008 Oct 3;283(40):27100-9. doi: 10.1074/jbc.M803923200. Epub 2008 Aug 6. J Biol Chem. 2008. PMID: 18684711 Free PMC article.
-
Low resolution structural study of two human HSP40 chaperones in solution. DJA1 from subfamily A and DJB4 from subfamily B have different quaternary structures.J Biol Chem. 2005 Apr 8;280(14):13671-81. doi: 10.1074/jbc.M408349200. Epub 2005 Jan 20. J Biol Chem. 2005. PMID: 15661747
-
The effect of mutating arginine-469 on the substrate binding and refolding activities of 70-kDa heat shock cognate protein.Arch Biochem Biophys. 2001 Feb 1;386(1):30-6. doi: 10.1006/abbi.2000.2176. Arch Biochem Biophys. 2001. PMID: 11360998
-
The Fink blueprint for Hsp70/Hsc70 molecular chaperones.Curr Protein Pept Sci. 2009 Oct;10(5):424-31. doi: 10.2174/138920309789352047. Curr Protein Pept Sci. 2009. PMID: 19538152 Review.
Cited by
-
Cochaperone binding to LYR motifs confers specificity of iron sulfur cluster delivery.Cell Metab. 2014 Mar 4;19(3):445-57. doi: 10.1016/j.cmet.2014.01.015. Cell Metab. 2014. PMID: 24606901 Free PMC article.
-
Hsp70/J-protein machinery from Glossina morsitans morsitans, vector of African trypanosomiasis.PLoS One. 2017 Sep 13;12(9):e0183858. doi: 10.1371/journal.pone.0183858. eCollection 2017. PLoS One. 2017. PMID: 28902917 Free PMC article.
-
Impact of heat shock transcription factor 1 on global gene expression profiles in cells which induce either cytoprotective or pro-apoptotic response following hyperthermia.BMC Genomics. 2013 Jul 8;14:456. doi: 10.1186/1471-2164-14-456. BMC Genomics. 2013. PMID: 23834426 Free PMC article.
-
The DNAJA2 substrate release mechanism is essential for chaperone-mediated folding.J Biol Chem. 2012 Dec 7;287(50):41939-54. doi: 10.1074/jbc.M112.413278. Epub 2012 Oct 22. J Biol Chem. 2012. PMID: 23091061 Free PMC article.
-
A cytosolic chaperone complex controls folding and degradation of type III CD38.J Biol Chem. 2019 Mar 15;294(11):4247-4258. doi: 10.1074/jbc.RA118.005844. Epub 2019 Jan 22. J Biol Chem. 2019. PMID: 30670591 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

