CK2 phosphorylation of Pdx-1 regulates its transcription factor activity
- PMID: 20339896
- PMCID: PMC11115922
- DOI: 10.1007/s00018-010-0348-0
CK2 phosphorylation of Pdx-1 regulates its transcription factor activity
Abstract
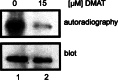
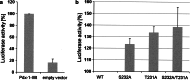
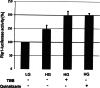
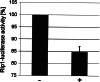
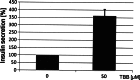
The duodenal homeobox-1 protein Pdx-1 is one of the regulators for the transcription of the insulin gene. Pdx-1 is a phosphoprotein, and there is increasing evidence for the regulation of some of its functions by phosphorylation. Here, we asked whether protein kinase CK2 might phosphorylate Pdx-1 and how this phosphorylation could be implicated in the functional regulation of Pdx-1. We used fragments of Pdx-1 as well as phosphorylation mutants for experiments with protein kinase CK2. Transactivation was measured by reporter assays using the insulin promoter. Our data showed that Pdx-1 is phosphorylated by protein kinase CK2 at amino acids thr(231) and ser(232), and this phosphorylation was implicated in the regulation of the transcription factor activity of Pdx-1. Furthermore, inhibition of protein kinase CK2 by specific inhibitors led to an elevated release of insulin from pancreatic beta-cells. Thus, these findings identify CK2 as a novel mediator of the insulin metabolism.
Figures
Similar articles
-
Functional interplay between the transcription factors USF1 and PDX-1 and protein kinase CK2 in pancreatic β-cells.Sci Rep. 2017 Nov 27;7(1):16367. doi: 10.1038/s41598-017-16590-0. Sci Rep. 2017. PMID: 29180680 Free PMC article.
-
Glucose regulates protein kinase CK2 in pancreatic β-cells and its interaction with PDX-1.Int J Biochem Cell Biol. 2013 Dec;45(12):2786-95. doi: 10.1016/j.biocel.2013.10.002. Epub 2013 Oct 12. Int J Biochem Cell Biol. 2013. PMID: 24126110
-
Per-Arnt-Sim kinase regulates pancreatic duodenal homeobox-1 protein stability via phosphorylation of glycogen synthase kinase 3β in pancreatic β-cells.J Biol Chem. 2013 Aug 23;288(34):24825-33. doi: 10.1074/jbc.M113.495945. Epub 2013 Jul 12. J Biol Chem. 2013. PMID: 23853095 Free PMC article.
-
Pancreatic duodenal homeobox factor-1 and diabetes mellitus type 2 (review).Int J Mol Med. 2008 Apr;21(4):399-404. Int J Mol Med. 2008. PMID: 18360684 Review.
-
PDX-1 functions as a master factor in the pancreas.Front Biosci. 2008 May 1;13:6406-20. doi: 10.2741/3162. Front Biosci. 2008. PMID: 18508668 Review.
Cited by
-
PDX1 is the cornerstone of pancreatic β-cell functions and identity.Front Mol Biosci. 2022 Dec 15;9:1091757. doi: 10.3389/fmolb.2022.1091757. eCollection 2022. Front Mol Biosci. 2022. PMID: 36589234 Free PMC article. Review.
-
Functional interplay between the transcription factors USF1 and PDX-1 and protein kinase CK2 in pancreatic β-cells.Sci Rep. 2017 Nov 27;7(1):16367. doi: 10.1038/s41598-017-16590-0. Sci Rep. 2017. PMID: 29180680 Free PMC article.
-
Targeting Protein Kinases to Protect Beta-Cell Function and Survival in Diabetes.Int J Mol Sci. 2024 Jun 11;25(12):6425. doi: 10.3390/ijms25126425. Int J Mol Sci. 2024. PMID: 38928130 Free PMC article. Review.
-
Protein Kinase CK2 Controls CaV2.1-Dependent Calcium Currents and Insulin Release in Pancreatic β-Cells.Int J Mol Sci. 2020 Jun 30;21(13):4668. doi: 10.3390/ijms21134668. Int J Mol Sci. 2020. PMID: 32630015 Free PMC article.
-
Adiponectin receptor 1 interacts with both subunits of protein kinase CK2.Mol Cell Biochem. 2011 Oct;356(1-2):185-9. doi: 10.1007/s11010-011-0941-z. Epub 2011 Jul 13. Mol Cell Biochem. 2011. PMID: 21750988
References
-
- Al-Quobaili F, Montenarh M. Pancreatic duodenal homeobox factor-1 and diabetes mellitus type 2. Int J Mol Med. 2008;21:399–404. - PubMed
-
- Amemiya-Kudo M, Oka J, Ide T, Matsuzaka T, Sone H, Yoshikawa T, Yahagi N, Ishibashi S, Osuga J, Yamada N, Murase T, Shimano H. Sterol regulatory element-binding proteins activate insulin gene promoter directly and indirectly through synergy with BETA2/E47. J Biol Chem. 2005;280:34577–34589. doi: 10.1074/jbc.M506718200. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

