Conservation of the TGFbeta/Labial homeobox signaling loop in endoderm-derived cells between Drosophila and mammals
- PMID: 20339309
- PMCID: PMC2865486
- DOI: 10.1159/000276895
Conservation of the TGFbeta/Labial homeobox signaling loop in endoderm-derived cells between Drosophila and mammals
Abstract
Background/aims: Midgut formation in Drosophila melanogaster is dependent upon the integrity of a signaling loop in the endoderm which requires the TGFbeta-related peptide, Decapentaplegic, and the Hox transcription factor, Labial. Interestingly, although Labial-like homeobox genes are present in mammals, their participation in endoderm morphogenesis is not clearly understood.
Methods: We report the cloning, expression, localization, TGFbeta inducibility, and biochemical properties of the mammalian Labial-like homeobox, HoxA1, in exocrine pancreatic cells that are embryologically derived from the gut endoderm.
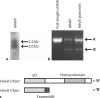
Results: HoxA1 is expressed in pancreatic cell populations as two alternatively spliced messages, encoding proteins that share their N-terminal domain, but either lack or include the homeobox at the C-terminus. Transcriptional regulatory assays demonstrate that the shared N-terminal domain behaves as a strong transcriptional activator in exocrine pancreatic cells. HoxA1 is an early response gene for TGFbeta(1) in pancreatic epithelial cell populations and HoxA1 protein co-localizes with TGFbeta(1) receptors in the embryonic pancreatic epithelium at a time when exocrine pancreatic morphogenesis occurs (days E16 and E17).
Conclusions: These results report a role for HoxA1 in linking TGFbeta-mediated signaling to gene expression in pancreatic epithelial cell populations, thus suggesting a high degree of conservation for a TGFbeta/labial signaling loop in endoderm-derived cells between Drosophila and mammals. and IAP.
Figures



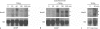
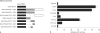
Similar articles
-
FoxK mediates TGF-beta signalling during midgut differentiation in flies.J Cell Biol. 2008 Dec 15;183(6):1049-60. doi: 10.1083/jcb.200808149. J Cell Biol. 2008. PMID: 19075113 Free PMC article.
-
Hox proteins coordinate peripodial decapentaplegic expression to direct adult head morphogenesis in Drosophila.Dev Biol. 2012 Sep 15;369(2):362-76. doi: 10.1016/j.ydbio.2012.07.012. Epub 2012 Jul 21. Dev Biol. 2012. PMID: 22824425 Free PMC article.
-
Drosophila endoderm development requires a novel homeobox gene which is a target of Wingless and Dpp signalling.Mech Dev. 1998 Dec;79(1-2):83-97. doi: 10.1016/s0925-4773(98)00172-5. Mech Dev. 1998. PMID: 10349623
-
The Cdx-1 and Cdx-2 homeobox genes in the intestine.Biochem Cell Biol. 1998;76(6):957-69. doi: 10.1139/o99-001. Biochem Cell Biol. 1998. PMID: 10392709 Review.
-
Cellular analysis of newly identified Hox downstream genes in Drosophila.Eur J Cell Biol. 2010 Feb-Mar;89(2-3):273-8. doi: 10.1016/j.ejcb.2009.11.012. Epub 2009 Dec 16. Eur J Cell Biol. 2010. PMID: 20018403 Review.
Cited by
-
Deciphering The Potential Role of Hox Genes in Pancreatic Cancer.Cancers (Basel). 2019 May 27;11(5):734. doi: 10.3390/cancers11050734. Cancers (Basel). 2019. PMID: 31137902 Free PMC article. Review.
-
LSD1-mediated enhancer silencing attenuates retinoic acid signalling during pancreatic endocrine cell development.Nat Commun. 2020 Apr 29;11(1):2082. doi: 10.1038/s41467-020-16017-x. Nat Commun. 2020. PMID: 32350257 Free PMC article.
References
-
- Manak JR, Scott MP. A class act: conservation of homeodomain protein functions. Dev Suppl. 1994:61–77. - PubMed
-
- Botas J. Control of morphogenesis and differentiation by HOM/HOX genes. Curr Opin Cell Biol. 1993;5:1015–1022. - PubMed
-
- Kappen C, Ruddle FH. Evolution of a regulatory gene family: HOM/HOX genes. Curr Opin Genet Dev. 1993;3:931–938. - PubMed
-
- Diederich RJ, Merrill VK, Pultz MA, Kaufman TC. Isolation, structure, and expression of labial, a homeotic gene of the antennapedia complex involved in Drosophila head development. Genes Dev. 1989;3:399–414. - PubMed
-
- Merrill VK, Diederich RJ, Turner FR, Kaufman TC. A genetic and developmental analysis of mutations in labial, a gene necessary for proper head formation in Drosophila melanogaster. Dev Biol. 1989;135:376–391. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

