Hormone binding and co-regulator binding to the glucocorticoid receptor are allosterically coupled
- PMID: 20335180
- PMCID: PMC2865338
- DOI: 10.1074/jbc.M110.108118
Hormone binding and co-regulator binding to the glucocorticoid receptor are allosterically coupled
Abstract
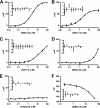
The glucocorticoid receptor initiates the cellular response to glucocorticoid steroid hormones in vertebrates. Co-regulator proteins dock to the receptor in response to hormone binding and potentiate the transcriptional activity of the receptor by modifying DNA and recruiting essential transcription factors like RNA polymerase II. Hormones and co-regulators bind at distinct sites in the ligand binding domain yet function cooperatively to mediate transcriptional control. This study reveals and quantifies energetic coupling between two binding sites using purified components. Using a library of peptides taken from co-regulator proteins, we determine the pattern of co-regulator binding to the glucocorticoid receptor ligand binding domain. We show that peptides from co-regulators differ in their effects on hormone binding and kinetics. Peptides from DAX1 and SRC1 bind with similar affinity, but DAX1 binding is coupled to hormone binding, and SRC1 is not. Mechanistic details of co-regulator binding and coupling to the hormone binding pocket are uncovered by analysis of properties endowed by mutation of a key residue in the allosteric network connecting the sites.
Figures

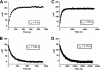

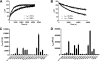
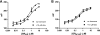

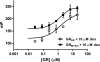

Similar articles
-
Glucocorticoid receptor ligand binding domain is sufficient for the modulation of glucocorticoid induction properties by homologous receptors, coactivator transcription intermediary factor 2, and Ubc9.Mol Endocrinol. 2005 Feb;19(2):290-311. doi: 10.1210/me.2004-0134. Epub 2004 Nov 11. Mol Endocrinol. 2005. PMID: 15539428
-
Distinct interaction of cortivazol with the ligand binding domain confers glucocorticoid receptor specificity: cortivazol is a specific ligand for the glucocorticoid receptor.J Biol Chem. 2002 Feb 15;277(7):5529-40. doi: 10.1074/jbc.M107946200. Epub 2001 Dec 10. J Biol Chem. 2002. PMID: 11741935
-
Specific DNA binding of Stat5, but not of glucocorticoid receptor, is required for their functional cooperation in the regulation of gene transcription.Mol Cell Biol. 1997 Nov;17(11):6708-16. doi: 10.1128/MCB.17.11.6708. Mol Cell Biol. 1997. PMID: 9343435 Free PMC article.
-
Glucocorticoid receptors.Monogr Endocrinol. 1979;12:49-77. doi: 10.1007/978-3-642-81265-1_3. Monogr Endocrinol. 1979. PMID: 386089 Review.
-
Mechanisms of glucocorticoid hormone action.J Steroid Biochem. 1984 Jan;20(1):77-88. doi: 10.1016/0022-4731(84)90192-4. J Steroid Biochem. 1984. PMID: 6368989 Review.
Cited by
-
Protein Ensembles: How Does Nature Harness Thermodynamic Fluctuations for Life? The Diverse Functional Roles of Conformational Ensembles in the Cell.Chem Rev. 2016 Jun 8;116(11):6516-51. doi: 10.1021/acs.chemrev.5b00562. Epub 2016 Jan 25. Chem Rev. 2016. PMID: 26807783 Free PMC article. Review.
-
Genetically tunable frustration controls allostery in an intrinsically disordered transcription factor.Elife. 2017 Oct 12;6:e30688. doi: 10.7554/eLife.30688. Elife. 2017. PMID: 29022880 Free PMC article.
-
Coupling of Conformational Transitions in the N-terminal Domain of the 51-kDa FK506-binding Protein (FKBP51) Near Its Site of Interaction with the Steroid Receptor Proteins.J Biol Chem. 2015 Jun 19;290(25):15746-15757. doi: 10.1074/jbc.M115.650655. Epub 2015 May 7. J Biol Chem. 2015. PMID: 25953903 Free PMC article.
-
Glucocorticoids, their uses, sexual dimorphisms, and diseases: new concepts, mechanisms, and discoveries.Physiol Rev. 2024 Jan 1;104(1):473-532. doi: 10.1152/physrev.00021.2023. Epub 2023 Sep 21. Physiol Rev. 2024. PMID: 37732829 Free PMC article. Review.
-
Rifampicin-independent interactions between the pregnane X receptor ligand binding domain and peptide fragments of coactivator and corepressor proteins.Biochemistry. 2012 Jan 10;51(1):19-31. doi: 10.1021/bi2011674. Epub 2011 Dec 20. Biochemistry. 2012. PMID: 22185585 Free PMC article.
References
-
- Katzenellenbogen B. S. (1980) Annu. Rev. Physiol. 42, 17–35 - PubMed
-
- Chrousos G. P., Kino T. (2007) Stress 10, 213–219 - PubMed
-
- Rhen T., Cidlowski J. A. (2005) N. Engl. J. Med. 353, 1711–1723 - PubMed
-
- Chrousos G. P., Charmandari E., Kino T. (2004) J. Clin. Endocrinol. Metab. 89, 563–564 - PubMed
-
- Pratt W. B., Dittmar K. D. (1998) Trends Endocrinol. Metab. 9, 244–252 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

