The complete plastid genome sequence of the secondarily nonphotosynthetic alga Cryptomonas paramecium: reduction, compaction, and accelerated evolutionary rate
- PMID: 20333213
- PMCID: PMC2839278
- DOI: 10.1093/gbe/evp047
The complete plastid genome sequence of the secondarily nonphotosynthetic alga Cryptomonas paramecium: reduction, compaction, and accelerated evolutionary rate
Abstract
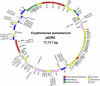
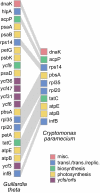
The cryptomonads are a group of unicellular algae that acquired photosynthesis through the engulfment of a red algal cell, a process called secondary endosymbiosis. Here, we present the complete plastid genome sequence of the secondarily nonphotosynthetic species Cryptomonas paramecium CCAP977/2a. The approximately 78 kilobase pair (Kbp) C. paramecium genome contains 82 predicted protein genes, 29 transfer RNA genes, and a single pseudogene (atpF). The C. paramecium plastid genome is approximately 50 Kbp smaller than those of the photosynthetic cryptomonads Guillardia theta and Rhodomonas salina; 71 genes present in the G. theta and/or R. salina plastid genomes are missing in C. paramecium. The pet, psa, and psb photosynthetic gene families are almost entirely absent. Interestingly, the ribosomal RNA operon, present as inverted repeats in most plastid genomes (including G. theta and R. salina), exists as a single copy in C. paramecium. The G + C content (38%) is higher in C. paramecium than in other cryptomonad plastid genomes, and C. paramecium plastid genes are characterized by significantly different codon usage patterns and increased evolutionary rates. The content and structure of the C. paramecium plastid genome provides insight into the changes associated with recent loss of photosynthesis in a predominantly photosynthetic group of algae and reveals features shared with the plastid genomes of other secondarily nonphotosynthetic eukaryotes.
Keywords: cryptomonads; genome reduction; photosynthesis; plastids; secondary endosymbiosis.
Figures


Similar articles
-
Complete nucleomorph genome sequence of the nonphotosynthetic alga Cryptomonas paramecium reveals a core nucleomorph gene set.Genome Biol Evol. 2011;3:44-54. doi: 10.1093/gbe/evq082. Epub 2010 Dec 8. Genome Biol Evol. 2011. PMID: 21147880 Free PMC article.
-
Comparative Plastid Genomics of Cryptomonas Species Reveals Fine-Scale Genomic Responses to Loss of Photosynthesis.Genome Biol Evol. 2020 Feb 1;12(2):3926-3937. doi: 10.1093/gbe/evaa001. Genome Biol Evol. 2020. PMID: 31922581 Free PMC article.
-
Nuclear genome sequence of the plastid-lacking cryptomonad Goniomonas avonlea provides insights into the evolution of secondary plastids.BMC Biol. 2018 Nov 28;16(1):137. doi: 10.1186/s12915-018-0593-5. BMC Biol. 2018. PMID: 30482201 Free PMC article.
-
A new scenario of plastid evolution: plastid primary endosymbiosis before the divergence of the "Plantae," emended.J Plant Res. 2005 Aug;118(4):247-55. doi: 10.1007/s10265-005-0219-1. Epub 2005 Jul 20. J Plant Res. 2005. PMID: 16032387 Review.
-
Photosynthetic eukaryotes unite: endosymbiosis connects the dots.Bioessays. 2004 Jan;26(1):50-60. doi: 10.1002/bies.10376. Bioessays. 2004. PMID: 14696040 Review.
Cited by
-
Nannochloropsis plastid and mitochondrial phylogenomes reveal organelle diversification mechanism and intragenus phylotyping strategy in microalgae.BMC Genomics. 2013 Aug 5;14:534. doi: 10.1186/1471-2164-14-534. BMC Genomics. 2013. PMID: 23915326 Free PMC article.
-
Comparative mitochondrial genomics of cryptophyte algae: gene shuffling and dynamic mobile genetic elements.BMC Genomics. 2018 Apr 20;19(1):275. doi: 10.1186/s12864-018-4626-9. BMC Genomics. 2018. PMID: 29678149 Free PMC article.
-
Reductive evolution of chloroplasts in non-photosynthetic plants, algae and protists.Curr Genet. 2018 Apr;64(2):365-387. doi: 10.1007/s00294-017-0761-0. Epub 2017 Oct 12. Curr Genet. 2018. PMID: 29026976 Review.
-
Highly Reduced Plastid Genomes of the Non-photosynthetic Dictyochophyceans Pteridomonas spp. (Ochrophyta, SAR) Are Retained for tRNA-Glu-Based Organellar Heme Biosynthesis.Front Plant Sci. 2020 Nov 27;11:602455. doi: 10.3389/fpls.2020.602455. eCollection 2020. Front Plant Sci. 2020. PMID: 33329672 Free PMC article.
-
The Plastid Genome of the Cryptomonad Teleaulax amphioxeia.PLoS One. 2015 Jun 5;10(6):e0129284. doi: 10.1371/journal.pone.0129284. eCollection 2015. PLoS One. 2015. PMID: 26047475 Free PMC article.
References
-
- Archibald JM. Nucleomorph genomes: structure, function, origin and evolution. Bioessays. 2007;29:392–402. - PubMed
-
- Archibald JM. The puzzle of plastid evolution. Curr Biol. 2009;19:R81–R88. - PubMed
-
- Barbrook AC, Howe CJ, Purton S. Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci. 2006;11:101–108. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

