Dynamic imaging of experimental Leishmania donovani-induced hepatic granulomas detects Kupffer cell-restricted antigen presentation to antigen-specific CD8 T cells
- PMID: 20300603
- PMCID: PMC2837408
- DOI: 10.1371/journal.ppat.1000805
Dynamic imaging of experimental Leishmania donovani-induced hepatic granulomas detects Kupffer cell-restricted antigen presentation to antigen-specific CD8 T cells
Abstract
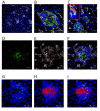
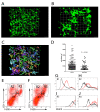
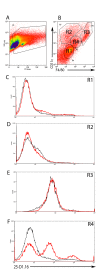
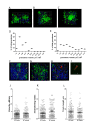
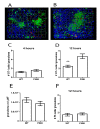
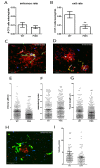
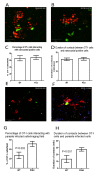
Kupffer cells (KCs) represent the major phagocytic population within the liver and provide an intracellular niche for the survival of a number of important human pathogens. Although KCs have been extensively studied in vitro, little is known of their in vivo response to infection and their capacity to directly interact with antigen-specific CD8(+) T cells. Here, using a combination of approaches including whole mount and thin section confocal microscopy, adoptive cell transfer and intra-vital 2-photon microscopy, we demonstrate that KCs represent the only detectable population of mononuclear phagocytes within granulomas induced by Leishmania donovani infection that are capable of presenting parasite-derived peptide to effector CD8(+) T cells. This restriction of antigen presentation to KCs within the Leishmania granuloma has important implications for the identification of new candidate vaccine antigens and for the design of novel immuno-therapeutic interventions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Hybrid cell vaccination resolves Leishmania donovani infection by eliciting a strong CD8+ cytotoxic T-lymphocyte response with concomitant suppression of interleukin-10 (IL-10) but not IL-4 or IL-13.Infect Immun. 2007 Dec;75(12):5956-66. doi: 10.1128/IAI.00944-07. Epub 2007 Oct 1. Infect Immun. 2007. PMID: 17908806 Free PMC article.
-
Histopathological and immunohistochemical characterisation of hepatic granulomas in Leishmania donovani-infected BALB/c mice: a time-course study.Parasit Vectors. 2018 Jan 31;11(1):73. doi: 10.1186/s13071-018-2624-z. Parasit Vectors. 2018. PMID: 29386047 Free PMC article.
-
Adoptive immunotherapy against experimental visceral leishmaniasis with CD8+ T cells requires the presence of cognate antigen.Infect Immun. 2006 Jan;74(1):773-6. doi: 10.1128/IAI.74.1.773-776.2006. Infect Immun. 2006. PMID: 16369038 Free PMC article.
-
Tissue granuloma structure-function in experimental visceral leishmaniasis.Int J Exp Pathol. 2001 Oct;82(5):249-67. doi: 10.1046/j.1365-2613.2001.00199.x. Int J Exp Pathol. 2001. PMID: 11703536 Free PMC article. Review.
-
Making an anti-amastigote vaccine for visceral leishmaniasis: rational, update and perspectives.Curr Opin Microbiol. 2012 Aug;15(4):476-85. doi: 10.1016/j.mib.2012.05.002. Epub 2012 Jun 13. Curr Opin Microbiol. 2012. PMID: 22698479 Review.
Cited by
-
Protective or Detrimental? Understanding the Role of Host Immunity in Leishmaniasis.Microorganisms. 2019 Dec 13;7(12):695. doi: 10.3390/microorganisms7120695. Microorganisms. 2019. PMID: 31847221 Free PMC article. Review.
-
A transcriptomic network identified in uninfected macrophages responding to inflammation controls intracellular pathogen survival.Cell Host Microbe. 2013 Sep 11;14(3):357-68. doi: 10.1016/j.chom.2013.08.004. Cell Host Microbe. 2013. PMID: 24034621 Free PMC article.
-
Pharmacodynamics and Biodistribution of Single-Dose Liposomal Amphotericin B at Different Stages of Experimental Visceral Leishmaniasis.Antimicrob Agents Chemother. 2017 Aug 24;61(9):e00497-17. doi: 10.1128/AAC.00497-17. Print 2017 Sep. Antimicrob Agents Chemother. 2017. PMID: 28630200 Free PMC article.
-
The Contribution of Immune Evasive Mechanisms to Parasite Persistence in Visceral Leishmaniasis.Front Immunol. 2016 Apr 22;7:153. doi: 10.3389/fimmu.2016.00153. eCollection 2016. Front Immunol. 2016. PMID: 27148272 Free PMC article. Review.
-
Granulomas in parasitic diseases: the good and the bad.Parasitol Res. 2020 Oct;119(10):3165-3180. doi: 10.1007/s00436-020-06841-x. Epub 2020 Aug 13. Parasitol Res. 2020. PMID: 32789534 Review.
References
-
- Tuijnman WB, Van Wichen DF, Schuurman HJ. Tissue distribution of human IgG Fc receptors CD16, CD32 and CD64: an immunohistochemical study. APMIS. 1993;101:319–329. - PubMed
-
- Seki E, Brenner DA. Toll-like receptors and adaptor molecules in liver disease: update. Hepatology. 2008;48:322–335. - PubMed
-
- Naito M, Hasegawa G, Ebe Y, Yamamoto T. Differentiation and function of Kupffer cells. Med Electron Microsc. 2004;37:16–28. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

