Single-molecule imaging brings Rad51 nucleoprotein filaments into focus
- PMID: 20299221
- PMCID: PMC2862779
- DOI: 10.1016/j.tcb.2010.02.004
Single-molecule imaging brings Rad51 nucleoprotein filaments into focus
Abstract
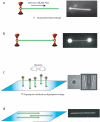
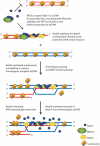
The Rad51 protein is essential for DNA repair by homologous recombination. After DNA damage, Rad51 localizes to nuclear foci that represent sites of DNA repair in vivo. In vitro, Rad51 self-assembles on single- or double-stranded DNA to form a nucleoprotein filament. Recently, the merging of innovative single-molecule techniques with ensemble methods has provided unique insights into the dynamic nature of this filament and its cellular function. The assembly and disassembly of Rad51 nucleoprotein filaments is seen to be regulated by recombination accessory proteins. In this regard, the BRC repeats of the BRCA2 protein were shown to modulate the DNA binding selectivity of Rad51. Furthermore, single-molecule studies explained the need for a DNA translocase, Rad54 protein, in the disassembly of Rad51 double-stranded DNA filaments.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures
Similar articles
-
Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function.Nucleic Acids Res. 2005 Jun 8;33(10):3292-302. doi: 10.1093/nar/gki640. Print 2005. Nucleic Acids Res. 2005. PMID: 15944450 Free PMC article.
-
Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats.Nat Struct Mol Biol. 2007 Jun;14(6):475-83. doi: 10.1038/nsmb1251. Nat Struct Mol Biol. 2007. PMID: 17515903 Free PMC article.
-
A novel function of Rad54 protein. Stabilization of the Rad51 nucleoprotein filament.J Biol Chem. 2003 Apr 18;278(16):14029-36. doi: 10.1074/jbc.M212779200. Epub 2003 Feb 3. J Biol Chem. 2003. PMID: 12566442
-
Single-molecule views on homologous recombination.Q Rev Biophys. 2013 Nov;46(4):323-48. doi: 10.1017/S0033583513000073. Epub 2013 Sep 9. Q Rev Biophys. 2013. PMID: 24016421 Review.
-
Insights into the control of RAD51 nucleoprotein filament dynamics from single-molecule studies.Curr Opin Genet Dev. 2021 Dec;71:182-187. doi: 10.1016/j.gde.2021.09.001. Epub 2021 Sep 24. Curr Opin Genet Dev. 2021. PMID: 34571340 Review.
Cited by
-
mRNA Expression and Methylation of the RAD51, ATM, ATR, BRCA1, and BRCA2 Genes in Gastric Adenocarcinoma.Biomark Insights. 2024 Jan 29;19:11772719231225206. doi: 10.1177/11772719231225206. eCollection 2024. Biomark Insights. 2024. PMID: 38293680 Free PMC article.
-
diRNA-Ago2-RAD51 complexes at double-strand break sites.Cell Res. 2014 May;24(5):511-2. doi: 10.1038/cr.2014.45. Epub 2014 Apr 11. Cell Res. 2014. PMID: 24722451 Free PMC article.
-
Single-molecule imaging of genome maintenance proteins encountering specific DNA sequences and structures.DNA Repair (Amst). 2023 Aug;128:103528. doi: 10.1016/j.dnarep.2023.103528. Epub 2023 Jun 24. DNA Repair (Amst). 2023. PMID: 37392578 Free PMC article. Review.
-
Immunomodulatory Effects of Radiotherapy.Int J Mol Sci. 2020 Oct 31;21(21):8151. doi: 10.3390/ijms21218151. Int J Mol Sci. 2020. PMID: 33142765 Free PMC article. Review.
-
Mechanisms governing the accessibility of DNA damage proteins to constitutive heterochromatin.Front Genet. 2022 Aug 26;13:876862. doi: 10.3389/fgene.2022.876862. eCollection 2022. Front Genet. 2022. PMID: 36092926 Free PMC article.
References
-
- Spies M, Kowalczykowski SC. Homologous recombination by RecBCD and RecF pathways. In: Higgins NP, editor. The Bacterial Chromosome. ASM Press; 2005. pp. 389–403.
-
- Lisby M, et al. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

