Cyclophilin B as a co-regulator of prolactin-induced gene expression and function in breast cancer cells
- PMID: 20237142
- PMCID: PMC2965652
- DOI: 10.1677/JME-09-0140
Cyclophilin B as a co-regulator of prolactin-induced gene expression and function in breast cancer cells
Abstract
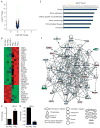
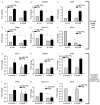
The effects of prolactin (PRL) during the pathogenesis of breast cancer are mediated in part though Stat5 activity enhanced by its interaction with its transcriptional inducer, the prolyl isomerase cyclophilin B (CypB). We have demonstrated that knockdown of CypB decreases cell growth, proliferation, and migration, and CypB expression is associated with malignant progression of breast cancer. In this study, we examined the effect of CypB knockdown on PRL signaling in breast cancer cells. CypB knockdown with two independent siRNAs was shown to impair PRL-induced reporter expression in breast cancer cell line. cDNA microarray analysis was performed on these cells to assess the effect of CypB reduction, and revealed a significant decrease in PRL-induced endogenous gene expression in two breast cancer cell lines. Parallel functional assays revealed corresponding alterations of both anchorage-independent cell growth and cell motility of breast cancer cells. Our results demonstrate that CypB expression levels significantly modulate PRL-induced function in breast cancer cells ultimately resulting in enhanced levels of PRL-responsive gene expression, cell growth, and migration. Given the increasingly appreciated role of PRL in the pathogenesis of breast cancer, the actions of CypB detailed here are of biological significance.
Conflict of interest statement
Figures
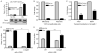
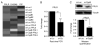

Similar articles
-
Role of cyclophilin B in prolactin signal transduction and nuclear retrotranslocation.Mol Endocrinol. 2000 Aug;14(8):1175-86. doi: 10.1210/mend.14.8.0508. Mol Endocrinol. 2000. PMID: 10935542
-
Expression of cyclophilin B is associated with malignant progression and regulation of genes implicated in the pathogenesis of breast cancer.Am J Pathol. 2009 Jan;174(1):297-308. doi: 10.2353/ajpath.2009.080753. Epub 2008 Dec 4. Am J Pathol. 2009. PMID: 19056847 Free PMC article.
-
Role of cyclophilins in somatolactogenic action.Ann N Y Acad Sci. 2000;917:514-21. doi: 10.1111/j.1749-6632.2000.tb05416.x. Ann N Y Acad Sci. 2000. PMID: 11268379 Review.
-
SOCS3 as a tumor suppressor in breast cancer cells, and its regulation by PRL.Int J Cancer. 2009 Apr 15;124(8):1756-66. doi: 10.1002/ijc.24172. Int J Cancer. 2009. PMID: 19115200
-
New mechanisms for PRLr action in breast cancer.Trends Endocrinol Metab. 2009 Jul;20(5):223-9. doi: 10.1016/j.tem.2009.03.001. Epub 2009 Jun 15. Trends Endocrinol Metab. 2009. PMID: 19535262 Review.
Cited by
-
Cyclophilin B supports Myc and mutant p53-dependent survival of glioblastoma multiforme cells.Cancer Res. 2014 Jan 15;74(2):484-96. doi: 10.1158/0008-5472.CAN-13-0771. Epub 2013 Nov 22. Cancer Res. 2014. PMID: 24272483 Free PMC article.
-
Cyclophilin B serum levels present variations across the menstrual cycle.Sci Rep. 2023 Jun 22;13(1):10139. doi: 10.1038/s41598-023-37322-7. Sci Rep. 2023. PMID: 37349369 Free PMC article.
-
Histone H1 and Chromosomal Protein HMGN2 Regulate Prolactin-induced STAT5 Transcription Factor Recruitment and Function in Breast Cancer Cells.J Biol Chem. 2017 Feb 10;292(6):2237-2254. doi: 10.1074/jbc.M116.764233. Epub 2016 Dec 29. J Biol Chem. 2017. PMID: 28035005 Free PMC article.
-
Cyclophilin B, a molecule chaperone, promotes adipogenesis in 3T3‑L1 preadipocytes via AKT/mTOR pathway.Int J Mol Med. 2023 Jan;51(1):6. doi: 10.3892/ijmm.2022.5209. Epub 2022 Dec 9. Int J Mol Med. 2023. PMID: 36484370 Free PMC article.
-
PPIB/Cyclophilin B expression associates with tumor progression and unfavorable survival in patients with pulmonary adenocarcinoma.Am J Cancer Res. 2024 Feb 25;14(2):917-930. doi: 10.62347/TYNU2341. eCollection 2024. Am J Cancer Res. 2024. PMID: 38455410 Free PMC article.
References
-
- Beck MT, Peirce SK, Chen WY. Regulation of bcl-2 gene expression in human breast cancer cells by prolactin and its antagonist, hPRL-G129R. Oncogene. 2002;21:5047–5055. - PubMed
-
- Breen EC, Tang K. Calcyclin (S100A6) regulates pulmonary fibroblast proliferation, morphology, and cytoskeletal organization in vitro. Journal of Cellular Biochemistry. 2003;88:848–854. - PubMed
-
- Brockman JL, Schroeder MD, Schuler LA. PRL activates the cyclin D1 promoter via the Jak2/Stat pathway. Molecular Endocrinology. 2002;16:774–784. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

