Basal core promoters control the equilibrium between negative cofactor 2 and preinitiation complexes in human cells
- PMID: 20230619
- PMCID: PMC2864573
- DOI: 10.1186/gb-2010-11-3-r33
Basal core promoters control the equilibrium between negative cofactor 2 and preinitiation complexes in human cells
Abstract
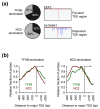
Background: The general transcription factor TFIIB and its antagonist negative cofactor 2 (NC2) are hallmarks of RNA polymerase II (RNAPII) transcription. Both factors bind TATA box-binding protein (TBP) at promoters in a mutually exclusive manner. Dissociation of NC2 is thought to be followed by TFIIB association and subsequent preinitiation complex formation. TFIIB dissociates upon RNAPII promoter clearance, thereby providing a specific measure for steady-state preinitiation complex levels. As yet, genome-scale promoter mapping of human TFIIB has not been reported. It thus remains elusive how human core promoters contribute to preinitiation complex formation in vivo.
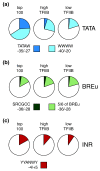
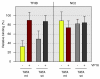
Results: We compare target genes of TFIIB and NC2 in human B cells and analyze associated core promoter architectures. TFIIB occupancy is positively correlated with gene expression, with the vast majority of promoters being GC-rich and lacking defined core promoter elements. TATA elements, but not the previously in vitro defined TFIIB recognition elements, are enriched in some 4 to 5% of the genes. NC2 binds to a highly related target gene set. Nonetheless, subpopulations show strong variations in factor ratios: whereas high TFIIB/NC2 ratios select for promoters with focused start sites and conserved core elements, high NC2/TFIIB ratios correlate to multiple start-site promoters lacking defined core elements.
Conclusions: TFIIB and NC2 are global players that occupy active genes. Preinitiation complex formation is independent of core elements at the majority of genes. TATA and TATA-like elements dictate TFIIB occupancy at a subset of genes. Biochemical data support a model in which preinitiation complex but not TBP-NC2 complex formation is regulated.
Figures


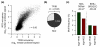

Similar articles
-
Global distribution of negative cofactor 2 subunit-alpha on human promoters.Proc Natl Acad Sci U S A. 2007 Jun 12;104(24):10000-5. doi: 10.1073/pnas.0703490104. Epub 2007 Jun 4. Proc Natl Acad Sci U S A. 2007. PMID: 17548813 Free PMC article.
-
Yeast NC2 associates with the RNA polymerase II preinitiation complex and selectively affects transcription in vivo.Mol Cell Biol. 2001 Apr;21(8):2736-42. doi: 10.1128/MCB.21.8.2736-2742.2001. Mol Cell Biol. 2001. PMID: 11283253 Free PMC article.
-
Efficient binding of NC2.TATA-binding protein to DNA in the absence of TATA.J Biol Chem. 2005 Feb 18;280(7):6222-30. doi: 10.1074/jbc.M406343200. Epub 2004 Nov 30. J Biol Chem. 2005. PMID: 15574413
-
Role of the TATA-box binding protein (TBP) and associated family members in transcription regulation.Gene. 2022 Jul 30;833:146581. doi: 10.1016/j.gene.2022.146581. Epub 2022 May 18. Gene. 2022. PMID: 35597524 Review.
-
TRF2: TRansForming the view of general transcription factors.Transcription. 2015;6(1):1-6. doi: 10.1080/21541264.2015.1004980. Epub 2015 Jan 14. Transcription. 2015. PMID: 25588059 Free PMC article. Review.
Cited by
-
Expression of the human endogenous retrovirus (HERV) group HML-2/HERV-K does not depend on canonical promoter elements but is regulated by transcription factors Sp1 and Sp3.J Virol. 2011 Apr;85(7):3436-48. doi: 10.1128/JVI.02539-10. Epub 2011 Jan 19. J Virol. 2011. PMID: 21248046 Free PMC article.
-
General transcription factor IIb overexpression and a potential link to proliferation in human hepatocellular carcinoma.Pathol Oncol Res. 2013 Apr;19(2):195-203. doi: 10.1007/s12253-012-9569-x. Epub 2012 Oct 5. Pathol Oncol Res. 2013. PMID: 23055019
-
Characterization of molecular and cellular functions of the cyclin-dependent kinase CDK9 using a novel specific inhibitor.Br J Pharmacol. 2014 Jan;171(1):55-68. doi: 10.1111/bph.12408. Br J Pharmacol. 2014. PMID: 24102143 Free PMC article.
-
Novel core promoter elements in the oomycete pathogen Phytophthora infestans and their influence on expression detected by genome-wide analysis.BMC Genomics. 2013 Feb 16;14:106. doi: 10.1186/1471-2164-14-106. BMC Genomics. 2013. PMID: 23414203 Free PMC article.
-
Focused transcription from the human CR2/CD21 core promoter is regulated by synergistic activity of TATA and Initiator elements in mature B cells.Cell Mol Immunol. 2016 Jan;13(1):119-31. doi: 10.1038/cmi.2014.138. Epub 2015 Feb 2. Cell Mol Immunol. 2016. PMID: 25640655 Free PMC article.
References
-
- Lifton RP, Goldberg ML, Karp RW, Hogness DS. The organization of the histone genes in Drosophila melanogaster : functional and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42:1047–1051. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

