Armadillo Repeat Containing 8alpha Binds to HRS and Promotes HRS Interaction with Ubiquitinated Proteins
- PMID: 20224683
- PMCID: PMC2835868
- DOI: 10.2174/1874091X01004010001
Armadillo Repeat Containing 8alpha Binds to HRS and Promotes HRS Interaction with Ubiquitinated Proteins
Abstract
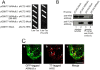
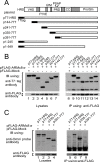
Recently, we reported that a complex with an essential role in the degradation of Fructose-1,6-bisphosphatase in yeast is well conserved in mammalian cells; we named this mammalian complex C-terminal to the Lissencephaly type-1-like homology (CTLH) complex. Although the function of the CTLH complex remains unclear, here we used yeast two-hybrid screening to isolate Hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) as a protein binding to a key component of CTLH complex, Armadillo repeat containing 8 (ARMc8) alpha. The association was confirmed by a yeast two-hybrid assay and a co-immunoprecipitation assay. The proline-rich domain of HRS was essential for the association. As demonstrated through immunofluorescence microscopy, ARMc8alpha co-localized with HRS. ARMc8alpha promoted the interaction of HRS with various ubiquitinated proteins through the ubiquitin-interacting motif. These findings suggest that HRS mediates protein endosomal trafficking partly through its interaction with ARMc8alpha.
Keywords: ARMc8α; FBPase; HRS; UIM.; monoubiquitination.
Figures
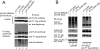
Similar articles
-
Proteasome-dependent degradation of alpha-catenin is regulated by interaction with ARMc8alpha.Biochem J. 2008 May 1;411(3):581-91. doi: 10.1042/BJ20071312. Biochem J. 2008. PMID: 18215130
-
RanBPM, Muskelin, p48EMLP, p44CTLH, and the armadillo-repeat proteins ARMC8alpha and ARMC8beta are components of the CTLH complex.Gene. 2007 Jul 15;396(2):236-47. doi: 10.1016/j.gene.2007.02.032. Epub 2007 Mar 24. Gene. 2007. PMID: 17467196
-
TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation.Proc Natl Acad Sci U S A. 2003 Jun 24;100(13):7626-31. doi: 10.1073/pnas.0932599100. Epub 2003 Jun 11. Proc Natl Acad Sci U S A. 2003. PMID: 12802020 Free PMC article.
-
Hrs and endocytic sorting of ubiquitinated membrane proteins.Cell Struct Funct. 2002 Dec;27(6):403-8. doi: 10.1247/csf.27.403. Cell Struct Funct. 2002. PMID: 12576633 Review.
-
Function of Hrs in endocytic trafficking and signalling.Biochem Soc Trans. 2001 Aug;29(Pt 4):472-5. doi: 10.1042/bst0290472. Biochem Soc Trans. 2001. PMID: 11498011 Review.
Cited by
-
Structural and Functional Insights into GID/CTLH E3 Ligase Complexes.Int J Mol Sci. 2022 May 24;23(11):5863. doi: 10.3390/ijms23115863. Int J Mol Sci. 2022. PMID: 35682545 Free PMC article. Review.
-
Molecular phylogeny of a RING E3 ubiquitin ligase, conserved in eukaryotic cells and dominated by homologous components, the muskelin/RanBPM/CTLH complex.PLoS One. 2013 Oct 15;8(10):e75217. doi: 10.1371/journal.pone.0075217. eCollection 2013. PLoS One. 2013. PMID: 24143168 Free PMC article.
-
Armc8 regulates the invasive ability of hepatocellular carcinoma through E-cadherin/catenin complex.Tumour Biol. 2016 Aug;37(8):11219-24. doi: 10.1007/s13277-016-5006-1. Epub 2016 Mar 4. Tumour Biol. 2016. PMID: 26944057
-
Studies of recombinant TWA1 reveal constitutive dimerization.Biosci Rep. 2017 Jan 13;37(1):BSR20160401. doi: 10.1042/BSR20160401. Print 2017 Feb 28. Biosci Rep. 2017. PMID: 27920276 Free PMC article.
-
RMND5 from Xenopus laevis is an E3 ubiquitin-ligase and functions in early embryonic forebrain development.PLoS One. 2015 Mar 20;10(3):e0120342. doi: 10.1371/journal.pone.0120342. eCollection 2015. PLoS One. 2015. PMID: 25793641 Free PMC article.
References
-
- Chiang H.L, Schekman R. Regulated import and degradation of a cytosolic protein in the yeast vacuole. Nature. 1991;350:313–318. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
