Cell type-specific gene expression differences in complex tissues
- PMID: 20208531
- PMCID: PMC3699332
- DOI: 10.1038/nmeth.1439
Cell type-specific gene expression differences in complex tissues
Abstract
We describe cell type-specific significance analysis of microarrays (csSAM) for analyzing differential gene expression for each cell type in a biological sample from microarray data and relative cell-type frequencies. First, we validated csSAM with predesigned mixtures and then applied it to whole-blood gene expression datasets from stable post-transplant kidney transplant recipients and those experiencing acute transplant rejection, which revealed hundreds of differentially expressed genes that were otherwise undetectable.
Figures
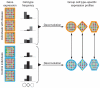

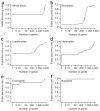
Comment in
-
Gene expression deconvolution in linear space.Nat Methods. 2011 Dec 28;9(1):8-9; author reply 9. doi: 10.1038/nmeth.1830. Nat Methods. 2011. PMID: 22205510 No abstract available.
Similar articles
-
A type I interferon signature characterizes chronic antibody-mediated rejection in kidney transplantation.J Pathol. 2015 Sep;237(1):72-84. doi: 10.1002/path.4553. Epub 2015 Jun 4. J Pathol. 2015. PMID: 25925804
-
Allo-specific immune response profiles indicative of acute rejection in kidney allografts using an in vitro lymphocyte culture-based model.Clin Exp Nephrol. 2018 Apr;22(2):465-473. doi: 10.1007/s10157-017-1469-7. Epub 2017 Aug 28. Clin Exp Nephrol. 2018. PMID: 28849286
-
Identification of T Cell-Mediated Vascular Rejection After Kidney Transplantation by the Combined Measurement of 5 Specific MicroRNAs in Blood.Transplantation. 2016 Apr;100(4):898-907. doi: 10.1097/TP.0000000000000873. Transplantation. 2016. PMID: 26444957
-
Novel insights into lung transplant rejection by microarray analysis.Proc Am Thorac Soc. 2007 Jan;4(1):44-51. doi: 10.1513/pats.200605-110JG. Proc Am Thorac Soc. 2007. PMID: 17202291 Free PMC article. Review.
-
Pre-processing of microarray data and analysis of differential expression.Methods Mol Biol. 2008;452:89-110. doi: 10.1007/978-1-60327-159-2_4. Methods Mol Biol. 2008. PMID: 18563370 Review.
Cited by
-
Identification of kidney cell types in scRNA-seq and snRNA-seq data using machine learning algorithms.Heliyon. 2024 Sep 27;10(19):e38567. doi: 10.1016/j.heliyon.2024.e38567. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39403515 Free PMC article.
-
Deconvolution from bulk gene expression by leveraging sample-wise and gene-wise similarities and single-cell RNA-Seq data.BMC Genomics. 2024 Sep 18;25(1):875. doi: 10.1186/s12864-024-10728-x. BMC Genomics. 2024. PMID: 39294558 Free PMC article.
-
BLEND: Probabilistic Cellular Deconvolution with Automated Reference Selection.bioRxiv [Preprint]. 2024 Aug 6:2024.08.02.606458. doi: 10.1101/2024.08.02.606458. bioRxiv. 2024. PMID: 39149243 Free PMC article. Preprint.
-
Investigating neonatal health risk variables through cell-type specific methylome-wide association studies.Clin Epigenetics. 2024 May 22;16(1):69. doi: 10.1186/s13148-024-01681-3. Clin Epigenetics. 2024. PMID: 38778395 Free PMC article.
-
CATD: a reproducible pipeline for selecting cell-type deconvolution methods across tissues.Bioinform Adv. 2024 Mar 23;4(1):vbae048. doi: 10.1093/bioadv/vbae048. eCollection 2024. Bioinform Adv. 2024. PMID: 38638280 Free PMC article.
References
Publication types
MeSH terms
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

