The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway
- PMID: 20192759
- PMCID: PMC3079308
- DOI: 10.1146/annurev.biochem.052308.093131
The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway
Abstract
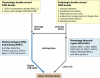
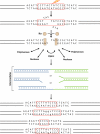
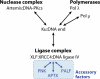
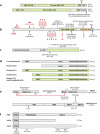
Double-strand DNA breaks are common events in eukaryotic cells, and there are two major pathways for repairing them: homologous recombination (HR) and nonhomologous DNA end joining (NHEJ). The various causes of double-strand breaks (DSBs) result in a diverse chemistry of DNA ends that must be repaired. Across NHEJ evolution, the enzymes of the NHEJ pathway exhibit a remarkable degree of structural tolerance in the range of DNA end substrate configurations upon which they can act. In vertebrate cells, the nuclease, DNA polymerases, and ligase of NHEJ are the most mechanistically flexible and multifunctional enzymes in each of their classes. Unlike repair pathways for more defined lesions, NHEJ repair enzymes act iteratively, act in any order, and can function independently of one another at each of the two DNA ends being joined. NHEJ is critical not only for the repair of pathologic DSBs as in chromosomal translocations, but also for the repair of physiologic DSBs created during variable (diversity) joining [V(D)J] recombination and class switch recombination (CSR). Therefore, patients lacking normal NHEJ are not only sensitive to ionizing radiation (IR), but also severely immunodeficient.
Figures
Similar articles
-
The mechanism of human nonhomologous DNA end joining.J Biol Chem. 2008 Jan 4;283(1):1-5. doi: 10.1074/jbc.R700039200. Epub 2007 Nov 12. J Biol Chem. 2008. PMID: 17999957 Review.
-
Analysis of chromatid-break-repair detects a homologous recombination to non-homologous end-joining switch with increasing load of DNA double-strand breaks.Mutat Res Genet Toxicol Environ Mutagen. 2021 Jul;867:503372. doi: 10.1016/j.mrgentox.2021.503372. Epub 2021 Jun 12. Mutat Res Genet Toxicol Environ Mutagen. 2021. PMID: 34266628
-
Mycobacterial nonhomologous end joining mediates mutagenic repair of chromosomal double-strand DNA breaks.J Bacteriol. 2007 Jul;189(14):5237-46. doi: 10.1128/JB.00332-07. Epub 2007 May 11. J Bacteriol. 2007. PMID: 17496093 Free PMC article.
-
PAXX and XLF DNA repair factors are functionally redundant in joining DNA breaks in a G1-arrested progenitor B-cell line.Proc Natl Acad Sci U S A. 2016 Sep 20;113(38):10619-24. doi: 10.1073/pnas.1611882113. Epub 2016 Sep 6. Proc Natl Acad Sci U S A. 2016. PMID: 27601633 Free PMC article.
-
Mechanisms of DNA double strand break repair and chromosome aberration formation.Cytogenet Genome Res. 2004;104(1-4):14-20. doi: 10.1159/000077461. Cytogenet Genome Res. 2004. PMID: 15162010 Review.
Cited by
-
Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells.Proc Natl Acad Sci U S A. 2013 May 7;110(19):7720-5. doi: 10.1073/pnas.1213431110. Epub 2013 Apr 22. Proc Natl Acad Sci U S A. 2013. PMID: 23610439 Free PMC article.
-
Biochemical and structural characterization of analogs of MRE11 breast cancer-associated mutant F237C.Sci Rep. 2021 Mar 29;11(1):7089. doi: 10.1038/s41598-021-86552-0. Sci Rep. 2021. PMID: 33782469 Free PMC article.
-
Modulation of DNA damage and repair pathways by human tumour viruses.Viruses. 2015 May 22;7(5):2542-91. doi: 10.3390/v7052542. Viruses. 2015. PMID: 26008701 Free PMC article. Review.
-
The recent advances in non-homologous end-joining through the lens of lymphocyte development.DNA Repair (Amst). 2020 Oct;94:102874. doi: 10.1016/j.dnarep.2020.102874. Epub 2020 Jun 25. DNA Repair (Amst). 2020. PMID: 32623318 Free PMC article. Review.
-
Bacterial DNA repair: recent insights into the mechanism of RecBCD, AddAB and AdnAB.Nat Rev Microbiol. 2013 Jan;11(1):9-13. doi: 10.1038/nrmicro2917. Epub 2012 Dec 3. Nat Rev Microbiol. 2013. PMID: 23202527 Review.
References
-
- Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–4. - PubMed
-
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–57. - PubMed
-
- Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

