Maternal control of early mouse development
- PMID: 20179092
- PMCID: PMC2834456
- DOI: 10.1242/dev.039487
Maternal control of early mouse development
Abstract
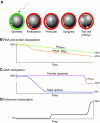
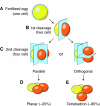
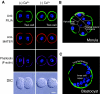
The hiatus between oocyte and embryonic gene transcription dictates a role for stored maternal factors in early mammalian development. Encoded by maternal-effect genes, these factors accumulate during oogenesis and enable the activation of the embryonic genome, the subsequent cleavage stages of embryogenesis and the initial establishment of embryonic cell lineages. Recent studies in mice have yielded new findings on the role of maternally provided proteins and multi-component complexes in preimplantation development. Nevertheless, significant gaps remain in our mechanistic understanding of the networks that regulate early mammalian embryogenesis, which provide an impetus and opportunities for future investigations.
Figures


Similar articles
-
[Crucial role of Polycomb proteins from maternal origin in mouse early embryonic development].Med Sci (Paris). 2012 Dec;28(12):1047-9. doi: 10.1051/medsci/20122812009. Epub 2012 Dec 21. Med Sci (Paris). 2012. PMID: 23290399 French. No abstract available.
-
What is the role of maternally provided Cdx2 mRNA in early mouse embryogenesis?Reprod Biomed Online. 2011 Jun;22(6):512-5. doi: 10.1016/j.rbmo.2011.02.003. Epub 2011 Feb 13. Reprod Biomed Online. 2011. PMID: 21498124
-
Post-translational regulation of the maternal-to-zygotic transition.Cell Mol Life Sci. 2018 May;75(10):1707-1722. doi: 10.1007/s00018-018-2750-y. Epub 2018 Feb 9. Cell Mol Life Sci. 2018. PMID: 29427077 Free PMC article. Review.
-
Degradation and translation of maternal mRNA for embryogenesis.Trends Genet. 2024 Mar;40(3):238-249. doi: 10.1016/j.tig.2023.12.008. Epub 2024 Jan 22. Trends Genet. 2024. PMID: 38262796 Review.
-
Molecular determinants of oocyte competence: potential functional role for maternal (oocyte-derived) follistatin in promoting bovine early embryogenesis.Endocrinology. 2009 May;150(5):2463-71. doi: 10.1210/en.2008-1574. Epub 2009 Jan 29. Endocrinology. 2009. PMID: 19179440
Cited by
-
The Maternal Effect Genes UTX and JMJD3 Play Contrasting Roles in Mus musculus Preimplantation Embryo Development.Sci Rep. 2016 Jul 7;6:26711. doi: 10.1038/srep26711. Sci Rep. 2016. PMID: 27384759 Free PMC article.
-
Low-input lipidomics reveals lipid metabolism remodelling during early mammalian embryo development.Nat Cell Biol. 2024 Feb;26(2):278-293. doi: 10.1038/s41556-023-01341-3. Epub 2024 Feb 1. Nat Cell Biol. 2024. PMID: 38302721
-
Preliminary crystallographic analysis of the N-terminal domain of FILIA, a protein essential for embryogenesis.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010 Sep 1;66(Pt 9):1111-4. doi: 10.1107/S1744309110031994. Epub 2010 Aug 28. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010. PMID: 20823540 Free PMC article.
-
LSM14B controls oocyte mRNA storage and stability to ensure female fertility.Cell Mol Life Sci. 2023 Aug 14;80(9):247. doi: 10.1007/s00018-023-04898-2. Cell Mol Life Sci. 2023. PMID: 37578641 Free PMC article.
-
Cloning of the Maternal Effector Gene org and Its Regulation by lncRNA ORG-AS in Chinese Tongue Sole (Cynoglossus semilaevis).Int J Mol Sci. 2022 Aug 3;23(15):8605. doi: 10.3390/ijms23158605. Int J Mol Sci. 2022. PMID: 35955739 Free PMC article.
References
-
- Adenot P. G., Mercier Y., Renard J. P., Thompson E. M. (1997). Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. aDevelopment 124, 4615-4625 - PubMed
-
- Alizadeh Z., Kageyama S., Aoki F. (2005). Degradation of maternal mRNA in mouse embryos: selective degradation of specific mRNAs after fertilization. Mol. Reprod. Dev. 72, 281-290 - PubMed
-
- Barton S. C., Surani M. A., Norris M. L. (1984). Role of paternal and maternal genomes in mouse development. Nature 311, 374-376 - PubMed
-
- Berryman M., Franck Z., Bretscher A. (1993). Ezrin is concentrated in the apical microvilli of a wide variety of epithelial cells whereas moesin is found primarily in endothelial cells. J. Cell Sci. 105, 1025-1043 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

