Xenopus meiotic microtubule-associated interactome
- PMID: 20174651
- PMCID: PMC2822853
- DOI: 10.1371/journal.pone.0009248
Xenopus meiotic microtubule-associated interactome
Abstract
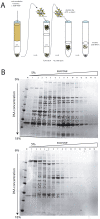
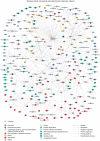
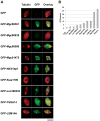
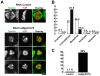
In metazoan oocytes the assembly of a microtubule-based spindle depends on the activity of a large number of accessory non-tubulin proteins, many of which remain unknown. In this work we isolated the microtubule-bound proteins from Xenopus eggs. Using mass spectrometry we identified 318 proteins, only 43 of which are known to bind microtubules. To integrate our results, we compiled for the first time a network of the meiotic microtubule-related interactome. The map reveals numerous interactions between spindle microtubules and the newly identified non-tubulin spindle components and highlights proteins absent from the mitotic spindle proteome. To validate newly identified spindle components, we expressed as GFP-fusions nine proteins identified by us and for first time demonstrated that Mgc68500, Loc398535, Nif3l1bp1/THOC7, LSM14A/RAP55A, TSGA14/CEP41, Mgc80361 and Mgc81475 are associated with spindles in egg extracts or in somatic cells. Furthermore, we showed that transfection of HeLa cells with siRNAs, corresponding to the human orthologue of Mgc81475 dramatically perturbs spindle formation in HeLa cells. These results show that our approach to the identification of the Xenopus microtubule-associated proteome yielded bona fide factors with a role in spindle assembly.
Conflict of interest statement
Figures

Similar articles
-
Visualizing and Analyzing Branching Microtubule Nucleation Using Meiotic Xenopus Egg Extracts and TIRF Microscopy.Methods Mol Biol. 2016;1413:77-85. doi: 10.1007/978-1-4939-3542-0_6. Methods Mol Biol. 2016. PMID: 27193844 Free PMC article.
-
The Xenopus TACC homologue, maskin, functions in mitotic spindle assembly.Mol Biol Cell. 2005 Jun;16(6):2836-47. doi: 10.1091/mbc.e04-10-0926. Epub 2005 Mar 23. Mol Biol Cell. 2005. PMID: 15788567 Free PMC article.
-
XMAP215 activity sets spindle length by controlling the total mass of spindle microtubules.Nat Cell Biol. 2013 Sep;15(9):1116-22. doi: 10.1038/ncb2834. Epub 2013 Aug 25. Nat Cell Biol. 2013. PMID: 23974040
-
Mechanism and function of poleward flux in Xenopus extract meiotic spindles.Philos Trans R Soc Lond B Biol Sci. 2005 Mar 29;360(1455):623-9. doi: 10.1098/rstb.2004.1616. Philos Trans R Soc Lond B Biol Sci. 2005. PMID: 15897184 Free PMC article. Review.
-
Mechanisms of Mitotic Spindle Assembly.Annu Rev Biochem. 2016 Jun 2;85:659-83. doi: 10.1146/annurev-biochem-060815-014528. Epub 2016 Apr 21. Annu Rev Biochem. 2016. PMID: 27145846 Free PMC article. Review.
Cited by
-
A microtubule-associated zinc finger protein, BuGZ, regulates mitotic chromosome alignment by ensuring Bub3 stability and kinetochore targeting.Dev Cell. 2014 Feb 10;28(3):268-81. doi: 10.1016/j.devcel.2013.12.013. Epub 2014 Jan 23. Dev Cell. 2014. PMID: 24462186 Free PMC article.
-
Proteomic Analysis of Microtubule Interacting Proteins over the Course of Xylem Tracheary Element Formation in Arabidopsis.Plant Cell. 2015 Oct;27(10):2709-26. doi: 10.1105/tpc.15.00314. Epub 2015 Oct 2. Plant Cell. 2015. PMID: 26432860 Free PMC article.
-
Purification and characterization of novel microtubule-associated proteins from Arabidopsis cell suspension cultures.Plant Physiol. 2013 Dec;163(4):1804-16. doi: 10.1104/pp.113.225607. Epub 2013 Oct 17. Plant Physiol. 2013. PMID: 24134884 Free PMC article.
-
Production of Xenopus tropicalis egg extracts to identify microtubule-associated RNAs.J Vis Exp. 2013 Jun 27;(76):50434. doi: 10.3791/50434. J Vis Exp. 2013. PMID: 23851481 Free PMC article.
-
Family-based exome sequencing and case-control analysis implicate CEP41 as an ASD gene.Transl Psychiatry. 2019 Jan 15;9(1):4. doi: 10.1038/s41398-018-0343-z. Transl Psychiatry. 2019. PMID: 30664616 Free PMC article.
References
-
- Castedo M, Kroemer G. [Mitotic catastrophe: a special case of apoptosis]. J Soc Biol. 2004;198:97–103. - PubMed
-
- Shi Q, King RW. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature. 2005;437:1038–1042. - PubMed
-
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. - PubMed
-
- Nicolaidis P, Petersen MB. Origin and mechanisms of non-disjunction in human autosomal trisomies. Hum Reprod. 1998;13:313–319. - PubMed
-
- Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

