Notch exhibits ligand bias and maneuvers stage-specific steering of neural differentiation in embryonic stem cells
- PMID: 20154142
- PMCID: PMC2849467
- DOI: 10.1128/MCB.01419-09
Notch exhibits ligand bias and maneuvers stage-specific steering of neural differentiation in embryonic stem cells
Abstract
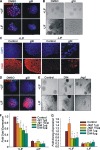
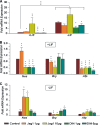
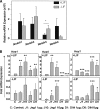
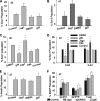
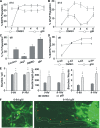
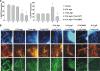
Notch dictates multiple developmental events, including stem cell maintenance and differentiation, through intercellular communication. However, its temporal influence during early development and, of particular interest, its regulation of binary fate decision at different stages during neurogenesis are among the least explored. Here, using an embryonic stem cell (ESC) model, we have deciphered Notch ligand preference during ESC commitment to different germ layers and determined the stage-specific temporal effect of Notch during neural differentiation. ESCs during maintenance remain impervious to Notch inhibition. However, Notch activation promotes differentiation even in the presence of leukemia inhibitory factor (LIF), displaying ligand preference-associated lineage discrimination, where Jagged-1 favors neural commitment and Delta-like-4 favors the mesoderm. This differential ligand action involves a combination of Notch receptors influencing specific downstream target gene expression. Though Notch activation during early neural differentiation specifically promotes neural stem cells or early neural progenitors and delays their maturation, its inhibition promotes late neural progenitors and expedites neurogenesis, with a preference for neurons over glia. However, gliogenesis is promoted upon Notch activation only when executed in combination with ciliary neurotrophic factor. Thus, our investigation underscores a multifaceted role of Notch, demonstrating the interdependency of ligand usage and lineage specification and Notch acting as a master switch, displaying stage-specific influence on neurogenesis.
Figures
Similar articles
-
Notch signaling is involved in neurogenic commitment of human periodontal ligament-derived mesenchymal stem cells.Stem Cells Dev. 2013 Apr 15;22(8):1220-31. doi: 10.1089/scd.2012.0430. Epub 2013 Feb 12. Stem Cells Dev. 2013. PMID: 23379739 Free PMC article.
-
Acheate-scute like 1 (Ascl1) is required for normal delta-like (Dll) gene expression and notch signaling during retinal development.Dev Dyn. 2009 Sep;238(9):2163-78. doi: 10.1002/dvdy.21848. Dev Dyn. 2009. PMID: 19191219 Free PMC article.
-
DLK1 promotes neurogenesis of human and mouse pluripotent stem cell-derived neural progenitors via modulating Notch and BMP signalling.Stem Cell Rev Rep. 2012 Jun;8(2):459-71. doi: 10.1007/s12015-011-9298-7. Stem Cell Rev Rep. 2012. PMID: 21761283
-
Cell and molecular biology of Notch.J Endocrinol. 2007 Sep;194(3):459-74. doi: 10.1677/JOE-07-0242. J Endocrinol. 2007. PMID: 17761886 Review.
-
Oscillatory Control of Notch Signaling in Development.Adv Exp Med Biol. 2018;1066:265-277. doi: 10.1007/978-3-319-89512-3_13. Adv Exp Med Biol. 2018. PMID: 30030831 Review.
Cited by
-
Notch signaling is involved in neurogenic commitment of human periodontal ligament-derived mesenchymal stem cells.Stem Cells Dev. 2013 Apr 15;22(8):1220-31. doi: 10.1089/scd.2012.0430. Epub 2013 Feb 12. Stem Cells Dev. 2013. PMID: 23379739 Free PMC article.
-
Notch3 signalling and vascular remodelling in pulmonary arterial hypertension.Clin Sci (Lond). 2019 Dec 20;133(24):2481-2498. doi: 10.1042/CS20190835. Clin Sci (Lond). 2019. PMID: 31868216 Free PMC article. Review.
-
Modeling Mammalian Commitment to the Neural Lineage Using Embryos and Embryonic Stem Cells.Front Physiol. 2019 Jul 11;10:705. doi: 10.3389/fphys.2019.00705. eCollection 2019. Front Physiol. 2019. PMID: 31354503 Free PMC article. Review.
-
High-mobility group nucleosomal binding domain 2 protects against microcephaly by maintaining global chromatin accessibility during corticogenesis.J Biol Chem. 2020 Jan 10;295(2):468-480. doi: 10.1074/jbc.RA119.010616. Epub 2019 Nov 7. J Biol Chem. 2020. PMID: 31699896 Free PMC article.
-
Spatial pattern dynamics of 3D stem cell loss of pluripotency via rules-based computational modeling.PLoS Comput Biol. 2013;9(3):e1002952. doi: 10.1371/journal.pcbi.1002952. Epub 2013 Mar 14. PLoS Comput Biol. 2013. PMID: 23516345 Free PMC article.
References
-
- Androutsellis-Theotokis, A., R. R. Leker, F. Soldner, D. J. Hoeppner, R. Ravin, S. W. Poser, M. A. Rueger, S. K. Bae, R. Kittappa, and R. D. McKay. 2006. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature 442:823-826. - PubMed
-
- Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770-776. - PubMed
-
- Bhattacharya, S., A. Das, K. Mallya, and I. Ahmad. 2008. CNTF-mediated signaling regulates neuronal versus glial differentiation of retinal stem cells/progenitors by concentration-dependent recruitment of MAPK and Jak-STAT pathways in conjunction with Notch signaling. Stem Cells 26:2611-2624. - PubMed
-
- Bonni, A., Y. Sun, M. Nadal-Vicens, A. Bhatt, D. Frank, I. Rozovsky, N. Stahl, G. Yancopoulos, and M. Greenberg. 1997. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science 278:477-483. - PubMed
-
- Chambers, I., D. Colby, M. Robertson, J. Nichols, S. Lee, S. Tweedie, and A. Smith. 2003. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113:643-655. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
