Roles of the lipid-binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy
- PMID: 20154084
- PMCID: PMC2857026
- DOI: 10.1074/jbc.M109.080374
Roles of the lipid-binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy
Abstract
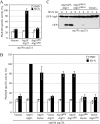
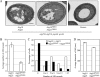
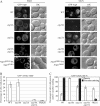
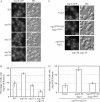
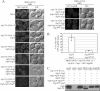
Atg18 and Atg21 are homologous WD-40 repeat proteins that bind phosphoinositides via a novel conserved Phe-Arg-Arg-Gly motif and function in autophagy-related pathways. Atg18 is required for the cytoplasm to vacuole targeting (Cvt) pathway and autophagy, whereas Atg21 is only required for the Cvt pathway. Currently, the functions of both proteins are poorly understood. Here, we examined the relationship between the phosphatidylinositol 3-phosphate (PtdIns(3)P)-binding abilities of Atg18 and Atg21 and autophagy by expressing variants of these proteins that have mutations in their phosphoinositide-binding motifs. Cells expressing PtdIns(3)P-binding mutants of both these proteins showed highly reduced autophagy. Furthermore, the localization of components of two related ubiquitin-like protein conjugation systems, Atg8 and Atg16, to the phagophore assembly site is affected. Consistent with the aberrant localization of the above Atg proteins, precursor Ape1, a cargo of the Cvt pathway and autophagy, is partially protease-sensitive in starvation conditions. This finding suggests a requirement for the PtdIns(3)P binding capability of Atg18 and Atg21 in efficient completion of the sequestering autophagic vesicles. Finally, using a multiple knock-out strain, we found that Atg18 and Atg21 facilitate the recruitment of Atg8-PE to the site of autophagosome formation and protect it from premature cleavage by Atg4, which represents a key aspect of post-translational autophagy regulation. Taken together, our results suggest that PtdIns(3)P binding by at least Atg18 or Atg21 is required for robust autophagic activity and that the PtdIns(3)P-binding motifs of Atg18 and Atg21 can compensate for one another in the recruitment of Atg components that are dependent on PtdIns(3)P for their phagophore assembly site association.
Figures

Similar articles
-
The relevance of the phosphatidylinositolphosphat-binding motif FRRGT of Atg18 and Atg21 for the Cvt pathway and autophagy.FEBS Lett. 2006 Aug 21;580(19):4632-8. doi: 10.1016/j.febslet.2006.07.041. Epub 2006 Jul 21. FEBS Lett. 2006. PMID: 16876790
-
Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy.Mol Biol Cell. 2004 Aug;15(8):3553-66. doi: 10.1091/mbc.e04-02-0147. Epub 2004 May 21. Mol Biol Cell. 2004. PMID: 15155809 Free PMC article.
-
Autophagic and non-autophagic functions of the Saccharomyces cerevisiae PROPPINs Atg18, Atg21 and Hsv2.Biol Chem. 2023 May 5;404(8-9):813-819. doi: 10.1515/hsz-2023-0126. Print 2023 Jul 26. Biol Chem. 2023. PMID: 37139661
-
Molecular machinery required for autophagy and the cytoplasm to vacuole targeting (Cvt) pathway in S. cerevisiae.Curr Opin Cell Biol. 2002 Aug;14(4):468-75. doi: 10.1016/s0955-0674(02)00343-5. Curr Opin Cell Biol. 2002. PMID: 12383798 Review.
-
Structural view on autophagosome formation.FEBS Lett. 2024 Jan;598(1):84-106. doi: 10.1002/1873-3468.14742. Epub 2023 Oct 5. FEBS Lett. 2024. PMID: 37758522 Review.
Cited by
-
Genome-wide association analysis identifies genetic loci associated with resistance to multiple antimalarials in Plasmodium falciparum from China-Myanmar border.Sci Rep. 2016 Oct 3;6:33891. doi: 10.1038/srep33891. Sci Rep. 2016. PMID: 27694982 Free PMC article.
-
Vps21 Directs the PI3K-PI(3)P-Atg21-Atg16 Module to Phagophores via Vps8 for Autophagy.Int J Mol Sci. 2022 Aug 23;23(17):9550. doi: 10.3390/ijms23179550. Int J Mol Sci. 2022. PMID: 36076954 Free PMC article.
-
Overexpression of MdATG18a in apple improves resistance to Diplocarpon mali infection by enhancing antioxidant activity and salicylic acid levels.Hortic Res. 2018 Nov 1;5:57. doi: 10.1038/s41438-018-0059-5. eCollection 2018. Hortic Res. 2018. PMID: 30393539 Free PMC article.
-
Activating Autophagy as a Therapeutic Strategy for Parkinson's Disease.CNS Drugs. 2018 Jan;32(1):1-11. doi: 10.1007/s40263-018-0497-5. CNS Drugs. 2018. PMID: 29492779 Review.
-
Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a β-propeller protein family.Proc Natl Acad Sci U S A. 2012 Jul 24;109(30):E2042-9. doi: 10.1073/pnas.1205128109. Epub 2012 Jul 2. Proc Natl Acad Sci U S A. 2012. PMID: 22753491 Free PMC article.
References
-
- Barth H., Meiling-Wesse K., Epple U. D., Thumm M. (2002) FEBS Lett. 512, 173–179 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

