Enrichment of homologs in insignificant BLAST hits by co-complex network alignment
- PMID: 20152020
- PMCID: PMC2836305
- DOI: 10.1186/1471-2105-11-86
Enrichment of homologs in insignificant BLAST hits by co-complex network alignment
Abstract
Background: Homology is a crucial concept in comparative genomics. The algorithm probably most widely used for homology detection in comparative genomics, is BLAST. Usually a stringent score cutoff is applied to distinguish putative homologs from possible false positive hits. As a consequence, some BLAST hits are discarded that are in fact homologous.
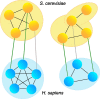
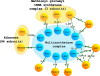
Results: Analogous to the use of the genomics context in genome alignments, we test whether conserved functional context can be used to select candidate homologs from insignificant BLAST hits. We make a co-complex network alignment between complex subunits in yeast and human and find that proteins with an insignificant BLAST hit that are part of homologous complexes, are likely to be homologous themselves. Further analysis of the distant homologs we recovered using the co-complex network alignment, shows that a large majority of these distant homologs are in fact ancient paralogs.
Conclusions: Our results show that, even though evolution takes place at the sequence and genome level, co-complex networks can be used as circumstantial evidence to improve confidence in the homology of distantly related sequences.
Figures


Similar articles
-
Identification of homologs in insignificant blast hits by exploiting extrinsic gene properties.BMC Bioinformatics. 2007 Sep 21;8:356. doi: 10.1186/1471-2105-8-356. BMC Bioinformatics. 2007. PMID: 17888146 Free PMC article.
-
Large-scale comparison of protein sequence alignment algorithms with structure alignments.Proteins. 2000 Jul 1;40(1):6-22. doi: 10.1002/(sici)1097-0134(20000701)40:1<6::aid-prot30>3.0.co;2-7. Proteins. 2000. PMID: 10813826
-
Detection of homologous proteins by an intermediate sequence search.Protein Sci. 2004 Jan;13(1):54-62. doi: 10.1110/ps.03335004. Protein Sci. 2004. PMID: 14691221 Free PMC article.
-
Finding homologs to nucleotide sequences using network BLAST searches.Curr Protoc Bioinformatics. 2002 Aug;Chapter 3:Unit 3.3. doi: 10.1002/0471250953.bi0303s00. Curr Protoc Bioinformatics. 2002. PMID: 18792938 Review.
-
An introduction to sequence similarity ("homology") searching.Curr Protoc Bioinformatics. 2013 Jun;Chapter 3:3.1.1-3.1.8. doi: 10.1002/0471250953.bi0301s42. Curr Protoc Bioinformatics. 2013. PMID: 23749753 Free PMC article. Review.
Cited by
-
Protein functional annotation of simultaneously improved stability, accuracy and false discovery rate achieved by a sequence-based deep learning.Brief Bioinform. 2020 Jul 15;21(4):1437-1447. doi: 10.1093/bib/bbz081. Brief Bioinform. 2020. PMID: 31504150 Free PMC article.
-
Phylometrics: a pipeline for inferring phylogenetic trees from a sequence relationship network perspective.BMC Bioinformatics. 2010 Oct 7;11 Suppl 6(Suppl 6):S18. doi: 10.1186/1471-2105-11-S6-S18. BMC Bioinformatics. 2010. PMID: 20946601 Free PMC article.
-
Extensive gene remodeling in the viral world: new evidence for nongradual evolution in the mobilome network.Genome Biol Evol. 2014 Aug 7;6(9):2195-205. doi: 10.1093/gbe/evu168. Genome Biol Evol. 2014. PMID: 25104113 Free PMC article.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

