Discrete subcellular partitioning of human retrotransposon RNAs despite a common mechanism of genome insertion
- PMID: 20147320
- PMCID: PMC2850619
- DOI: 10.1093/hmg/ddq048
Discrete subcellular partitioning of human retrotransposon RNAs despite a common mechanism of genome insertion
Abstract
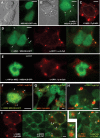
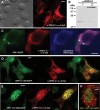
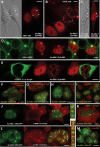
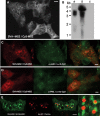
Despite the immense significance retrotransposons have had for genome evolution much about their biology is unknown, including the processes of forming their ribonucleoprotein (RNP) particles and transporting them about the cell. Suppression of retrotransposon expression, together with the presence of retrotransposon sequence within numerous mRNAs, makes tracking endogenous L1 RNP particles in cells problematic. We overcame these difficulties by assaying in living and fixed cells tagged-RNPs generated from constructs expressing retrotransposition-competent L1s. In this way, we demonstrate for the first time the subcellular colocalization of L1 RNA and proteins ORF1p and ORF2p, and show their targeting together to cytoplasmic foci. Foci are often associated with markers of cytoplasmic stress granules. Furthermore, mutation analyses reveal that ORF1p can direct L1 RNP distribution within the cell. We also assayed RNA localization of the non-autonomous retrotransposons Alu and SVA. Despite a requirement for the L1 integration machinery, each manifests unique features of subcellular RNA distribution. In nuclei Alu RNA forms small round foci partially associated with marker proteins for coiled bodies, suborganelles involved in the processing of non-coding RNAs. SVA RNA patterning is distinctive, being cytoplasmic but without prominent foci and concentrated in large nuclear aggregates that often ring nucleoli. Such variability predicts significant differences in the life cycles of these elements.
Figures

Similar articles
-
Characterization of LINE-1 ribonucleoprotein particles.PLoS Genet. 2010 Oct 7;6(10):e1001150. doi: 10.1371/journal.pgen.1001150. PLoS Genet. 2010. PMID: 20949108 Free PMC article.
-
Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition.Hum Mol Genet. 2005 Nov 1;14(21):3237-48. doi: 10.1093/hmg/ddi354. Epub 2005 Sep 23. Hum Mol Genet. 2005. PMID: 16183655
-
Enrichment of processed pseudogene transcripts in L1-ribonucleoprotein particles.Hum Mol Genet. 2013 Sep 15;22(18):3730-48. doi: 10.1093/hmg/ddt225. Epub 2013 May 21. Hum Mol Genet. 2013. PMID: 23696454 Free PMC article.
-
An update on post-transcriptional regulation of retrotransposons.FEBS Lett. 2023 Feb;597(3):380-406. doi: 10.1002/1873-3468.14551. Epub 2022 Dec 12. FEBS Lett. 2023. PMID: 36460901 Review.
-
Protein-nucleic acid interactions of LINE-1 ORF1p.Semin Cell Dev Biol. 2019 Feb;86:140-149. doi: 10.1016/j.semcdb.2018.03.019. Epub 2018 Mar 31. Semin Cell Dev Biol. 2019. PMID: 29596909 Free PMC article. Review.
Cited by
-
mRNA Targeting, Transport and Local Translation in Eukaryotic Cells: From the Classical View to a Diversity of New Concepts.Mol Biol. 2021;55(4):507-537. doi: 10.1134/S0026893321030080. Epub 2021 May 30. Mol Biol. 2021. PMID: 34092811 Free PMC article.
-
LINE-1 mRNA 3' end dynamics shape its biology and retrotransposition potential.Nucleic Acids Res. 2024 Apr 12;52(6):3327-3345. doi: 10.1093/nar/gkad1251. Nucleic Acids Res. 2024. PMID: 38197223 Free PMC article.
-
LINEs, SINEs and other retroelements: do birds of a feather flock together?Front Biosci (Landmark Ed). 2012 Jan 1;17(4):1345-61. doi: 10.2741/3991. Front Biosci (Landmark Ed). 2012. PMID: 22201808 Free PMC article. Review.
-
Spliced integrated retrotransposed element (SpIRE) formation in the human genome.PLoS Biol. 2018 Mar 5;16(3):e2003067. doi: 10.1371/journal.pbio.2003067. eCollection 2018 Mar. PLoS Biol. 2018. PMID: 29505568 Free PMC article.
-
HIV-2 genomic RNA accumulates in stress granules in the absence of active translation.Nucleic Acids Res. 2014 Nov 10;42(20):12861-75. doi: 10.1093/nar/gku1017. Epub 2014 Oct 28. Nucleic Acids Res. 2014. PMID: 25352557 Free PMC article.
References
-
- Temin H.M., Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226:1211–1213. - PubMed
-
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed

