Kinetic mechanistic studies of wild-type leucine-rich repeat kinase 2: characterization of the kinase and GTPase activities
- PMID: 20146535
- PMCID: PMC2897705
- DOI: 10.1021/bi901851y
Kinetic mechanistic studies of wild-type leucine-rich repeat kinase 2: characterization of the kinase and GTPase activities
Abstract
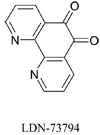
Recent studies have identified mutations in the leucine-rich repeat kinase2 gene (LRRK2) in the most common familial forms and some sporadic forms of Parkinson's disease (PD). LRRK2 is a large and complex protein that possesses kinase and GTPase activities. Some LRRK2 mutants enhance kinase activity and possibly contribute to PD through a toxic gain-of-function mechanism. Given the role of LRRK2 in the pathogenesis of PD, understanding the kinetic mechanism of its two enzymatic properties is critical for the discovery of inhibitors of LRRK2 kinase that would be therapeutically useful in treating PD. In this report, by using LRRK2 protein purified from murine brain, first we characterize kinetic mechanisms for the LRRK2-catalyzed phosphorylation of two peptide substrates: PLK-derived peptide (PLK-peptide) and LRRKtide. We found that LRRK2 follows a rapid equilibrium random mechanism for the phosphorylation of PLK-peptide with either ATP or PLK-peptide being the first substrate binding to the enzyme, as evidenced by initial velocity and inhibition mechanism studies with nucleotide analogues AMP and AMP-PNP, product ADP, and an analogue of the peptide substrate. The binding of the first substrate has no effect on the binding affinity of the second substrate. Identical mechanistic conclusions were drawn when LRRKtide was the phosphoryl acceptor. Next, we characterize the GTPase activity of LRRK2 with a k(cat) of 0.2 +/- 0.02 s(-1) and a K(m) of 210 +/- 29 microM. A SKIE of 0.97 +/- 0.04 was measured on k(cat) for the GTPase activity of LRRK2 in a D(2)O molar fraction of 0.86 and suggested that the product dissociation step is rate-limiting, of the steps governed by k(cat) in the LRRK2-catalyzed GTP hydrolysis. Surprisingly, binding of GTP, GDP, or GMP has no effect on kinase activity, although GMP and GDP inhibit the GTPase activity. Finally, we have identified compound LDN-73794 through screen of LRRK2 kinase inhibitors. Our study revealed that this compound is a competitive inhibitor of the binding of ATP and inhibits the kinase activity without affecting the GTPase activity.
Figures







Similar articles
-
Leucine-rich repeat kinase 2 (LRRK2)/PARK8 possesses GTPase activity that is altered in familial Parkinson's disease R1441C/G mutants.J Neurochem. 2007 Oct;103(1):238-47. doi: 10.1111/j.1471-4159.2007.04743.x. Epub 2007 Jul 10. J Neurochem. 2007. PMID: 17623048 Free PMC article.
-
Kinetic, mechanistic, and structural modeling studies of truncated wild-type leucine-rich repeat kinase 2 and the G2019S mutant.Biochemistry. 2011 Nov 1;50(43):9399-408. doi: 10.1021/bi201173d. Epub 2011 Oct 7. Biochemistry. 2011. PMID: 21961647 Free PMC article.
-
Development of a mechanism-based high-throughput screen assay for leucine-rich repeat kinase 2--discovery of LRRK2 inhibitors.Anal Biochem. 2010 Sep 15;404(2):186-92. doi: 10.1016/j.ab.2010.05.033. Epub 2010 Jun 2. Anal Biochem. 2010. PMID: 20566370 Free PMC article.
-
Contribution of GTPase activity to LRRK2-associated Parkinson disease.Small GTPases. 2013 Jul-Sep;4(3):164-70. doi: 10.4161/sgtp.25130. Epub 2013 Jun 10. Small GTPases. 2013. PMID: 24025585 Free PMC article. Review.
-
LRRK2 GTPase dysfunction in the pathogenesis of Parkinson's disease.Biochem Soc Trans. 2012 Oct;40(5):1074-9. doi: 10.1042/BST20120093. Biochem Soc Trans. 2012. PMID: 22988868 Free PMC article. Review.
Cited by
-
Membrane localization of LRRK2 is associated with increased formation of the highly active LRRK2 dimer and changes in its phosphorylation.Biochemistry. 2010 Jul 6;49(26):5511-23. doi: 10.1021/bi100157u. Biochemistry. 2010. PMID: 20515039 Free PMC article.
-
Identification and characterization of a leucine-rich repeat kinase 2 (LRRK2) consensus phosphorylation motif.PLoS One. 2010 Oct 27;5(10):e13672. doi: 10.1371/journal.pone.0013672. PLoS One. 2010. PMID: 21060682 Free PMC article.
-
LRRK2, a puzzling protein: insights into Parkinson's disease pathogenesis.Exp Neurol. 2014 Nov;261:206-16. doi: 10.1016/j.expneurol.2014.05.025. Epub 2014 Jun 4. Exp Neurol. 2014. PMID: 24907399 Free PMC article. Review.
-
Is inhibition of kinase activity the only therapeutic strategy for LRRK2-associated Parkinson's disease?BMC Med. 2012 Feb 23;10:20. doi: 10.1186/1741-7015-10-20. BMC Med. 2012. PMID: 22361010 Free PMC article. Review.
-
The dual enzyme LRRK2 hydrolyzes GTP in both its GTPase and kinase domains in vitro.Biochim Biophys Acta Proteins Proteom. 2017 Mar;1865(3):274-280. doi: 10.1016/j.bbapap.2016.12.001. Epub 2016 Dec 8. Biochim Biophys Acta Proteins Proteom. 2017. PMID: 27939437 Free PMC article.
References
-
- Moore DJ, West AB, Dawson VL, Dawson TM. Molecular pathophysiology of Parkinson's disease. Annu. Rev. Neurosci. 2005;28:57–87. - PubMed
-
- Paisán-Ruíz C, Jain S, Evans EW, Gilks WP, Simón J, van der Brug M, López de Munain A, Aparicio S, Gil AM, Khan N, Johnson J, Martinez JR, Nicholl D, Carrera IM, Pena AS, de Silva R, Lees A, Martí-Massó JF, Pérez-Tur J, Wood NW, Singleton AB. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. - PubMed
-
- Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, Stoessl AJ, Pfeiffer RF, Patenge N, Carbajal IC, Vieregge P, Asmus F, Müller-Myhsok B, Dickson DW, Meitinger T, Strom TM, Wszolek ZK, Gasser T. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. - PubMed
-
- Berg D, Schweitzer K, Leitner P, Zimprich A, Lichtner P, Belcredi P, Brussel T, Schulte C, Maass S, Nagele T. Type and frequency of mutations in the LRRK2 gene in familial and sporadic Parkinson's disease. Brain. 2005;128:3000–3011. - PubMed
-
- Khan NL, Jain S, Lynch JM, Pavese N, Abou-Sleiman P, Holton JL, Healy DG, Gilks WP, Sweeney MG, Ganguly M, Gibbons V, Gandhi S, Vaughan J, Eunson LH, Katzenschlager R, Gayton J, Lennox G, Revesz T, Nicholl D, Bhatia KP, Quinn N, Brooks D, Lees AJ, Davis MB, Piccini P, Singleton AB, Wood NW. Mutations in the gene LRRK2 encoding dardarin (PARK8) cause familial Parkinson's disease: clinical, pathological, olfactory and functional imaging and genetic data. Brain. 2005;128:2786–2796. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

