Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes
- PMID: 20144761
- PMCID: PMC2821818
- DOI: 10.1016/j.cell.2009.12.054
Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes
Abstract
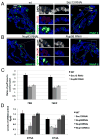
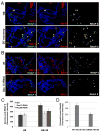
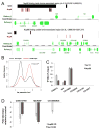
Nuclear pore complexes have recently been shown to play roles in gene activation; however their potential involvement in metazoan transcription remains unclear. Here we show that the nucleoporins Sec13, Nup98, and Nup88, as well as a group of FG-repeat nucleoporins, bind to the Drosophila genome at functionally distinct loci that often do not represent nuclear envelope contact sites. Whereas Nup88 localizes to silent loci, Sec13, Nup98, and a subset of FG-repeat nucleoporins bind to developmentally regulated genes undergoing transcription induction. Strikingly, RNAi-mediated knockdown of intranuclear Sec13 and Nup98 specifically inhibits transcription of their target genes and prevents efficient reactivation of transcription after heat shock, suggesting an essential role of NPC components in regulating complex gene expression programs of multicellular organisms.
2010 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm.Cell. 2010 Feb 5;140(3):360-71. doi: 10.1016/j.cell.2010.01.011. Cell. 2010. PMID: 20144760
-
Characterization of genome-nucleoporin interactions in Drosophila links chromatin insulators to the nuclear pore complex.Cell Cycle. 2010 Dec 15;9(24):4812-7. doi: 10.4161/cc.9.24.14328. Epub 2010 Dec 15. Cell Cycle. 2010. PMID: 21150273
-
Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome.PLoS Genet. 2010 Feb 12;6(2):e1000846. doi: 10.1371/journal.pgen.1000846. PLoS Genet. 2010. PMID: 20174442 Free PMC article.
-
The Nuclear Pore Complex in Cell Type-Specific Chromatin Structure and Gene Regulation.Trends Genet. 2019 Aug;35(8):579-588. doi: 10.1016/j.tig.2019.05.006. Epub 2019 Jun 15. Trends Genet. 2019. PMID: 31213386 Review.
-
Nucleoplasmic Nup98 controls gene expression by regulating a DExH/D-box protein.Nucleus. 2018 Jan 1;9(1):1-8. doi: 10.1080/19491034.2017.1364826. Epub 2017 Sep 21. Nucleus. 2018. PMID: 28934014 Free PMC article. Review.
Cited by
-
Nuclear envelope and genome interactions in cell fate.Front Genet. 2015 Mar 19;6:95. doi: 10.3389/fgene.2015.00095. eCollection 2015. Front Genet. 2015. PMID: 25852741 Free PMC article. Review.
-
An Overview of Genome Organization and How We Got There: from FISH to Hi-C.Microbiol Mol Biol Rev. 2015 Sep;79(3):347-72. doi: 10.1128/MMBR.00006-15. Microbiol Mol Biol Rev. 2015. PMID: 26223848 Free PMC article. Review.
-
Isolation of chromatin DNA tightly bound to the nuclear envelope of HeLa cells.J Membr Biol. 2012 Nov;245(11):683-90. doi: 10.1007/s00232-012-9437-3. Epub 2012 May 30. J Membr Biol. 2012. PMID: 22644390
-
"Off-pore" nucleoporins relocalize heterochromatic breaks through phase separation.bioRxiv [Preprint]. 2024 Jul 18:2023.12.07.570729. doi: 10.1101/2023.12.07.570729. bioRxiv. 2024. PMID: 39071440 Free PMC article. Preprint.
-
A genome-wide 3C-method for characterizing the three-dimensional architectures of genomes.Methods. 2012 Nov;58(3):277-88. doi: 10.1016/j.ymeth.2012.06.018. Epub 2012 Jul 6. Methods. 2012. PMID: 22776363 Free PMC article. Review.
References
-
- Agudo D, Gomez-Esquer F, Martinez-Arribas F, Nunez-Villar MJ, Pollan M, Schneider J. Nup88 mRNA overexpression is associated with high aggressiveness of breast cancer. Int J Cancer. 2004;109:717–720. - PubMed
-
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. - PubMed
-
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007a;450:683–694. - PubMed
-
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007b;450:695–701. - PubMed
-
- Andres AJ, Thummel CS. Methods for quantitative analysis of transcription in larvae and prepupae. Methods Cell Biol. 1994;44:565–573. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

