Functional residues on the surface of the N-terminal domain of yeast Pms1
- PMID: 20138591
- PMCID: PMC2856611
- DOI: 10.1016/j.dnarep.2010.01.010
Functional residues on the surface of the N-terminal domain of yeast Pms1
Abstract
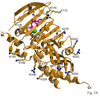
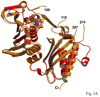
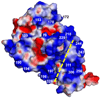
Saccharomyces cerevisiae MutLalpha is a heterodimer of Mlh1 and Pms1 that participates in DNA mismatch repair (MMR). Both proteins have weakly conserved C-terminal regions (CTDs), with the CTD of Pms1 harboring an essential endonuclease activity. These proteins also have conserved N-terminal domains (NTDs) that bind and hydrolyze ATP and bind to DNA. To better understand Pms1 functions and potential interactions with DNA and/or other proteins, we solved the 2.5A crystal structure of yeast Pms1 (yPms1) NTD. The structure is similar to the homologous NTDs of Escherichia coli MutL and human PMS2, including the site involved in ATP binding and hydrolysis. The structure reveals a number of conserved, positively charged surface residues that do not interact with other residues in the NTD and are therefore candidates for interactions with DNA, with the CTD and/or with other proteins. When these were replaced with glutamate, several replacements resulted in yeast strains with elevated mutation rates. Two replacements also resulted in NTDs with decreased DNA binding affinity in vitro, suggesting that these residues contribute to DNA binding that is important for mismatch repair. Elevated mutation rates also resulted from surface residue replacements that did not affect DNA binding, suggesting that these conserved residues serve other functions, possibly involving interactions with other MMR proteins.
Published by Elsevier B.V.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures










Similar articles
-
Molecular basis of the dual role of the Mlh1-Mlh3 endonuclease in MMR and in meiotic crossover formation.Proc Natl Acad Sci U S A. 2021 Jun 8;118(23):e2022704118. doi: 10.1073/pnas.2022704118. Proc Natl Acad Sci U S A. 2021. PMID: 34088835 Free PMC article.
-
Mapping the dimer interface in the C-terminal domains of the yeast MLH1-PMS1 heterodimer.Biochemistry. 2006 Dec 26;45(51):15458-67. doi: 10.1021/bi061392a. Epub 2006 Dec 8. Biochemistry. 2006. PMID: 17176067
-
Differential ATP binding and intrinsic ATP hydrolysis by amino-terminal domains of the yeast Mlh1 and Pms1 proteins.J Biol Chem. 2002 Feb 1;277(5):3673-9. doi: 10.1074/jbc.M106120200. Epub 2001 Nov 20. J Biol Chem. 2002. PMID: 11717305
-
DNA mismatch repair and mutation avoidance pathways.J Cell Physiol. 2002 Apr;191(1):28-41. doi: 10.1002/jcp.10077. J Cell Physiol. 2002. PMID: 11920679 Review.
-
DNA binding properties of the yeast Msh2-Msh6 and Mlh1-Pms1 heterodimers.Biol Chem. 2002 Jun;383(6):969-75. doi: 10.1515/BC.2002.103. Biol Chem. 2002. PMID: 12222686 Review.
Cited by
-
The unstructured linker arms of Mlh1-Pms1 are important for interactions with DNA during mismatch repair.J Mol Biol. 2012 Sep 14;422(2):192-203. doi: 10.1016/j.jmb.2012.05.030. Epub 2012 May 30. J Mol Biol. 2012. PMID: 22659005 Free PMC article.
-
DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae.Genetics. 2013 Apr;193(4):1025-64. doi: 10.1534/genetics.112.145219. Genetics. 2013. PMID: 23547164 Free PMC article. Review.
-
Dynamic human MutSα-MutLα complexes compact mismatched DNA.Proc Natl Acad Sci U S A. 2020 Jul 14;117(28):16302-16312. doi: 10.1073/pnas.1918519117. Epub 2020 Jun 25. Proc Natl Acad Sci U S A. 2020. PMID: 32586954 Free PMC article.
-
Experimental exchange of paralogous domains in the MLH family provides evidence of sub-functionalization after gene duplication.G3 (Bethesda). 2021 Jun 17;11(6):jkab111. doi: 10.1093/g3journal/jkab111. G3 (Bethesda). 2021. PMID: 33871573 Free PMC article.
-
Small-angle X-ray scattering analysis reveals the ATP-bound monomeric state of the ATPase domain from the homodimeric MutL endonuclease, a GHKL phosphotransferase superfamily protein.Extremophiles. 2015 May;19(3):643-56. doi: 10.1007/s00792-015-0745-2. Epub 2015 Mar 26. Extremophiles. 2015. PMID: 25809295
References
-
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. - PubMed
-
- Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. - PubMed
-
- Harfe BD, Jinks-Robertson S. DNA mismatch repair and genetic instability. Annu Rev Genet. 2000;34:359–399. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

