Cell-nonautonomous regulation of C. elegans germ cell death by kri-1
- PMID: 20137949
- PMCID: PMC2829125
- DOI: 10.1016/j.cub.2009.12.032
Cell-nonautonomous regulation of C. elegans germ cell death by kri-1
Abstract
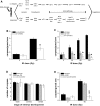
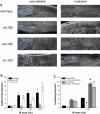
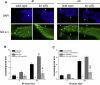
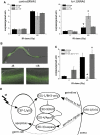
Programmed cell death (or apoptosis) is an evolutionarily conserved, genetically controlled suicide mechanism for cells that, when deregulated, can lead to developmental defects, cancers, and degenerative diseases. In C. elegans, DNA damage induces germ cell death by signaling through cep-1/p53, ultimately leading to the activation of CED-3/caspase. It has been hypothesized that the major regulatory events controlling cell death occur by cell-autonomous mechanisms, that is, within the dying cell. In support of this, genetic studies in C. elegans have shown that the core apoptosis pathway genes ced-4/APAF-1 and ced-3/caspase are required in cells fated to die. However, it is not known whether the upstream signals that activate apoptosis function in a cell-autonomous manner. Here we show that kri-1, an ortholog of KRIT1/CCM1, which is mutated in the human neurovascular disease cerebral cavernous malformation, is required to activate DNA damage-dependent cell death independently of cep-1/p53. Interestingly, we find that kri-1 regulates cell death in a cell-nonautonomous manner, revealing a novel regulatory role for nondying cells in eliciting cell death in response to DNA damage.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures
Similar articles
-
C. elegans CEP-1/p53 and BEC-1 are involved in DNA repair.PLoS One. 2014 Feb 20;9(2):e88828. doi: 10.1371/journal.pone.0088828. eCollection 2014. PLoS One. 2014. PMID: 24586407 Free PMC article.
-
C. elegans ced-13 can promote apoptosis and is induced in response to DNA damage.Cell Death Differ. 2005 Feb;12(2):153-61. doi: 10.1038/sj.cdd.4401539. Cell Death Differ. 2005. PMID: 15605074
-
Regulation of Caenorhabditis elegans p53/CEP-1-dependent germ cell apoptosis by Ras/MAPK signaling.PLoS Genet. 2011 Aug;7(8):e1002238. doi: 10.1371/journal.pgen.1002238. Epub 2011 Aug 25. PLoS Genet. 2011. PMID: 21901106 Free PMC article.
-
Death and more: DNA damage response pathways in the nematode C. elegans.Cell Death Differ. 2004 Jan;11(1):21-8. doi: 10.1038/sj.cdd.4401340. Cell Death Differ. 2004. PMID: 14685168 Review.
-
Methods for studying the DNA damage response in the Caenorhabdatis elegans germ line.Methods Cell Biol. 2012;107:321-52. doi: 10.1016/B978-0-12-394620-1.00011-4. Methods Cell Biol. 2012. PMID: 22226529 Review.
Cited by
-
High-content imaging-based pooled CRISPR screens in mammalian cells.J Cell Biol. 2021 Feb 1;220(2):e202008158. doi: 10.1083/jcb.202008158. J Cell Biol. 2021. PMID: 33465779 Free PMC article.
-
Pro-crossover factors regulate damage-dependent apoptosis in the Caenorhabditis elegans germ line.Cell Death Differ. 2013 Sep;20(9):1209-18. doi: 10.1038/cdd.2013.68. Epub 2013 Jul 5. Cell Death Differ. 2013. PMID: 23832114 Free PMC article.
-
Somatic gonad sheath cells and Eph receptor signaling promote germ-cell death in C. elegans.Cell Death Differ. 2012 Jun;19(6):1080-9. doi: 10.1038/cdd.2011.192. Epub 2012 Jan 13. Cell Death Differ. 2012. PMID: 22240896 Free PMC article.
-
Noncanonical cell death in the nematode Caenorhabditis elegans.Methods Enzymol. 2014;545:157-80. doi: 10.1016/B978-0-12-801430-1.00007-X. Methods Enzymol. 2014. PMID: 25065890 Free PMC article. Review.
-
Systematic pharmacological screens uncover novel pathways involved in cerebral cavernous malformations.EMBO Mol Med. 2018 Oct;10(10):e9155. doi: 10.15252/emmm.201809155. EMBO Mol Med. 2018. PMID: 30181117 Free PMC article.
References
-
- Ellis H.M., Horvitz H.R. Genetic control of programmed cell death in the nematode C. elegans. Cell. 1986;44:817–829. - PubMed
-
- Hipfner D.R., Cohen S.M. Connecting proliferation and apoptosis in development and disease. Nat. Rev. Mol. Cell Biol. 2004;5:805–815. - PubMed
-
- Ahmed S., Hodgkin J. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature. 2000;403:159–164. - PubMed
-
- Gartner A., Milstein S., Ahmed S., Hodgkin J., Hengartner M.O. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell. 2000;5:435–443. - PubMed
-
- Hofmann E.R., Milstein S., Boulton S.J., Ye M., Hofmann J.J., Stergiou L., Gartner A., Vidal M., Hengartner M.O. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr. Biol. 2002;12:1908–1918. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

