RAD51D protects against MLH1-dependent cytotoxic responses to O(6)-methylguanine
- PMID: 20133210
- PMCID: PMC2858319
- DOI: 10.1016/j.dnarep.2010.01.009
RAD51D protects against MLH1-dependent cytotoxic responses to O(6)-methylguanine
Abstract
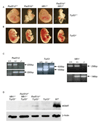
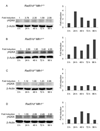
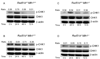
S(N)1-type methylating agents generate O(6)-methyl guanine (O(6)-meG), which is a potently mutagenic, toxic, and recombinogenic DNA adduct. Recognition of O(6)-meG:T mismatches by mismatch repair (MMR) causes sister chromatid exchanges, which are representative of homologous recombination (HR) events. Although the MMR-dependent mutagenicity and toxicity caused by O(6)-meG has been studied, the mechanisms of recombination induced by O(6)-meG are poorly understood. To explore the HR and MMR genetic interactions in mammals, we used the Rad51d and Mlh1 mouse models. Ablation of Mlh1 did not appreciably influence the developmental phenotypes conferred by the absence of Rad51d. Mouse embryonic fibroblasts (MEFs) deficient in Rad51d can only proliferate in p53-deficient background. Therefore, Rad51d(-/-)Mlh1(-/-)Trp53(-/-) MEFs with a combined deficiency of HR and MMR were generated and comparisons between MLH1 and RAD51D status were made. To our knowledge, these MEFs are the first mammalian model system for combined HR and MMR defects. Rad51d-deficient MEFs were 5.3-fold sensitive to N-methyl-N'-nitro-N-nitrosoguanidine (MNNG) compared to the Rad51d-proficient MEFs. A pronounced G2/M arrest in Rad51d-deficient cells was accompanied by an accumulation of gamma-H2AX and apoptosis. Mlh1-deficient MEFs were resistant to MNNG and showed no G2/M arrest or apoptosis at the doses used. Importantly, loss of Mlh1 alleviated sensitivity of Rad51d-deficient cells to MNNG, in addition to reducing gamma-H2AX, G2/M arrest and apoptosis. Collectively, the data support the hypothesis that MMR-dependent sensitization of HR-deficient cells is specific for O(6)-meG and suggest that HR resolves DNA intermediates created by MMR recognition of O(6)-meG:T. This study provides insight into recombinogenic mechanisms of carcinogenesis and chemotherapy resulting from O(6)-meG adducts.
2010 Elsevier B.V. All rights reserved.
Figures




Similar articles
-
Brca2/Xrcc2 dependent HR, but not NHEJ, is required for protection against O(6)-methylguanine triggered apoptosis, DSBs and chromosomal aberrations by a process leading to SCEs.DNA Repair (Amst). 2009 Jan 1;8(1):72-86. doi: 10.1016/j.dnarep.2008.09.003. Epub 2008 Oct 21. DNA Repair (Amst). 2009. PMID: 18840549
-
MMR/c-Abl-dependent activation of ING2/p73alpha signaling regulates the cell death response to N-methyl-N'-nitro-N-nitrosoguanidine.Exp Cell Res. 2009 Nov 1;315(18):3163-75. doi: 10.1016/j.yexcr.2009.09.010. Epub 2009 Sep 17. Exp Cell Res. 2009. PMID: 19766113 Free PMC article.
-
The base excision repair enzyme MED1 mediates DNA damage response to antitumor drugs and is associated with mismatch repair system integrity.Proc Natl Acad Sci U S A. 2003 Dec 9;100(25):15071-6. doi: 10.1073/pnas.2334585100. Epub 2003 Nov 12. Proc Natl Acad Sci U S A. 2003. PMID: 14614141 Free PMC article.
-
Signalling cell cycle arrest and cell death through the MMR System.Carcinogenesis. 2006 Apr;27(4):682-92. doi: 10.1093/carcin/bgi298. Epub 2005 Dec 6. Carcinogenesis. 2006. PMID: 16332722 Review.
-
Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification.J Med Genet. 2012 Mar;49(3):151-7. doi: 10.1136/jmedgenet-2011-100714. J Med Genet. 2012. PMID: 22368298 Review.
Cited by
-
XPA Enhances Temozolomide Resistance of Glioblastoma Cells by Promoting Nucleotide Excision Repair.Cell Transplant. 2022 Jan-Dec;31:9636897221092778. doi: 10.1177/09636897221092778. Cell Transplant. 2022. PMID: 35536165 Free PMC article.
-
XAB2 promotes Ku eviction from single-ended DNA double-strand breaks independently of the ATM kinase.Nucleic Acids Res. 2021 Sep 27;49(17):9906-9925. doi: 10.1093/nar/gkab785. Nucleic Acids Res. 2021. PMID: 34500463 Free PMC article.
-
Exonuclease 1 (Exo1) is required for activating response to S(N)1 DNA methylating agents.DNA Repair (Amst). 2012 Dec 1;11(12):951-64. doi: 10.1016/j.dnarep.2012.09.004. Epub 2012 Oct 11. DNA Repair (Amst). 2012. PMID: 23062884 Free PMC article.
-
DNA mismatch repair and the DNA damage response.DNA Repair (Amst). 2016 Feb;38:94-101. doi: 10.1016/j.dnarep.2015.11.019. Epub 2015 Dec 2. DNA Repair (Amst). 2016. PMID: 26704428 Free PMC article. Review.
-
Multifaceted roles of alkyltransferase and related proteins in DNA repair, DNA damage, resistance to chemotherapy, and research tools.Chem Res Toxicol. 2011 May 16;24(5):618-39. doi: 10.1021/tx200031q. Epub 2011 Apr 28. Chem Res Toxicol. 2011. PMID: 21466232 Free PMC article. Review.
References
-
- Margison GP, Santibanez Koref MF, Povey AC. Mechanisms of carcinogenicity/chemotherapy by O6-methylguanine. Mutagenesis. 2002;17:483–487. - PubMed
-
- Sedgwick B. Repairing DNA-methylation damage. Nat Rev Mol Cell Biol. 2004;5:148–157. - PubMed
-
- Kaina B, Christmann M, Naumann S, Roos WP. MGMT: Key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair. 2007;6:1079–1099. - PubMed
-
- Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004;4:296–307. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

