Sirtuin chemical mechanisms
- PMID: 20132909
- PMCID: PMC2886189
- DOI: 10.1016/j.bbapap.2010.01.021
Sirtuin chemical mechanisms
Abstract
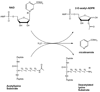
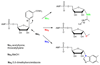
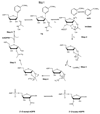
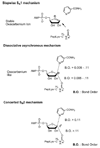
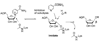
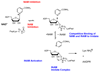
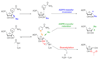
Sirtuins are ancient proteins widely distributed in all lifeforms of earth. These proteins are universally able to bind NAD(+), and activate it to effect ADP-ribosylation of cellular nucleophiles. The most commonly observed sirtuin reaction is the ADP-ribosylation of acetyllysine, which leads to NAD(+)-dependent deacetylation. Other types of ADP-ribosylation have also been observed, including protein ADP-ribosylation, NAD(+) solvolysis and ADP-ribosyltransfer to 5,6-dimethylbenzimidazole, a reaction involved in eubacterial cobalamin biosynthesis. This review broadly surveys the chemistries and chemical mechanisms of these enzymes.
Copyright 2010 Elsevier B.V. All rights reserved.
Figures

Similar articles
-
SIR2: the biochemical mechanism of NAD(+)-dependent protein deacetylation and ADP-ribosyl enzyme intermediates.Curr Med Chem. 2004 Apr;11(7):807-26. doi: 10.2174/0929867043455675. Curr Med Chem. 2004. PMID: 15078167 Review.
-
Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases.Proc Natl Acad Sci U S A. 2004 Jun 8;101(23):8563-8. doi: 10.1073/pnas.0401057101. Epub 2004 May 18. Proc Natl Acad Sci U S A. 2004. PMID: 15150415 Free PMC article.
-
Plasmodium falciparum Sir2 is an NAD+-dependent deacetylase and an acetyllysine-dependent and acetyllysine-independent NAD+ glycohydrolase.Biochemistry. 2008 Sep 23;47(38):10227-39. doi: 10.1021/bi800767t. Epub 2008 Aug 26. Biochemistry. 2008. PMID: 18729382 Free PMC article.
-
Sirtuins: NAD(+)-dependent deacetylase mechanism and regulation.Curr Opin Chem Biol. 2012 Dec;16(5-6):535-43. doi: 10.1016/j.cbpa.2012.10.003. Epub 2012 Oct 23. Curr Opin Chem Biol. 2012. PMID: 23102634 Review.
-
Investigating the ADP-ribosyltransferase activity of sirtuins with NAD analogues and 32P-NAD.Biochemistry. 2009 Apr 7;48(13):2878-90. doi: 10.1021/bi802093g. Biochemistry. 2009. PMID: 19220062
Cited by
-
Sirtuin Deacetylation Mechanism and Catalytic Role of the Dynamic Cofactor Binding Loop.J Phys Chem Lett. 2013 Feb 7;4(3):491-495. doi: 10.1021/jz302015s. J Phys Chem Lett. 2013. PMID: 23585919 Free PMC article.
-
Acylation of Biomolecules in Prokaryotes: a Widespread Strategy for the Control of Biological Function and Metabolic Stress.Microbiol Mol Biol Rev. 2015 Sep;79(3):321-46. doi: 10.1128/MMBR.00020-15. Epub 2015 Jul 15. Microbiol Mol Biol Rev. 2015. PMID: 26179745 Free PMC article. Review.
-
Gatekeepers of chromatin: Small metabolites elicit big changes in gene expression.Trends Biochem Sci. 2012 Nov;37(11):477-83. doi: 10.1016/j.tibs.2012.07.008. Epub 2012 Sep 1. Trends Biochem Sci. 2012. PMID: 22944281 Free PMC article. Review.
-
Sirtuin activators and inhibitors: Promises, achievements, and challenges.Pharmacol Ther. 2018 Aug;188:140-154. doi: 10.1016/j.pharmthera.2018.03.004. Epub 2018 Mar 22. Pharmacol Ther. 2018. PMID: 29577959 Free PMC article. Review.
-
Eight Kinetically Stable but Thermodynamically Activated Molecules that Power Cell Metabolism.Chem Rev. 2018 Feb 28;118(4):1460-1494. doi: 10.1021/acs.chemrev.7b00510. Epub 2017 Dec 22. Chem Rev. 2018. PMID: 29272116 Free PMC article. Review.
References
-
- Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–465. - PubMed
-
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. - PubMed
-
- Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat Rev Genet. 2007;8:835–844. - PubMed
-
- Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

