Force spectroscopy and fluorescence microscopy of dsDNA-YOYO-1 complexes: implications for the structure of dsDNA in the overstretching region
- PMID: 20129944
- PMCID: PMC2879515
- DOI: 10.1093/nar/gkq034
Force spectroscopy and fluorescence microscopy of dsDNA-YOYO-1 complexes: implications for the structure of dsDNA in the overstretching region
Abstract
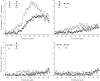
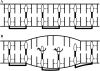
When individual dsDNA molecules are stretched beyond their B-form contour length, they reveal a structural transition in which the molecule extends 1.7 times its contour length. The nature of this transition is still a subject of debate. In the first model, the DNA helix unwinds and combined with the tilting of the base pairs (which remain intact), results in a stretched form of DNA (also known as S-DNA). In the second model the base pairs break resulting effectively in two single-strands, which is referred to as force-induced melting. Here a combination of optical tweezers force spectroscopy with fluorescence microscopy was used to study the structure of dsDNA in the overstretching regime. When dsDNA was stretched in the presence of 10 nM YOYO-1 an initial increase in total fluorescence intensity of the dye-DNA complex was observed and at an extension where the dsDNA started to overstretch the fluorescence intensity leveled off and ultimately decreased when stretched further into the overstretching region. Simultaneous force spectroscopy and fluorescence polarization microscopy revealed that the orientation of dye molecules did not change significantly in the overstretching region (78.0 degrees +/- 3.2 degrees). These results presented here clearly suggest that, the structure of overstretched dsDNA can be explained accurately by force induced melting.
Figures

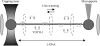



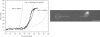
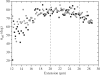
Similar articles
-
Interaction of oxazole yellow dyes with DNA studied with hybrid optical tweezers and fluorescence microscopy.Biophys J. 2009 Aug 5;97(3):835-43. doi: 10.1016/j.bpj.2009.05.024. Biophys J. 2009. PMID: 19651041 Free PMC article.
-
The kinetics of YOYO-1 intercalation into single molecules of double-stranded DNA.Biochem Biophys Res Commun. 2010 Dec 10;403(2):225-9. doi: 10.1016/j.bbrc.2010.11.015. Epub 2010 Nov 10. Biochem Biophys Res Commun. 2010. PMID: 21073861
-
1H NMR studies of the bis-intercalation of a homodimeric oxazole yellow dye in DNA oligonucleotides.J Biomol Struct Dyn. 1998 Oct;16(2):205-22. doi: 10.1080/07391102.1998.10508240. J Biomol Struct Dyn. 1998. PMID: 9833661
-
Denaturation transition of stretched DNA.Biochem Soc Trans. 2013 Apr;41(2):639-45. doi: 10.1042/BST20120298. Biochem Soc Trans. 2013. PMID: 23514169 Review.
-
Single-Molecule Methods for Investigating the Double-Stranded DNA Bendability.Mol Cells. 2022 Jan 31;45(1):33-40. doi: 10.14348/molcells.2021.0182. Mol Cells. 2022. PMID: 34470919 Free PMC article. Review.
Cited by
-
The Statistical Segment Length of DNA: Opportunities for Biomechanical Modeling in Polymer Physics and Next-Generation Genomics.J Biomech Eng. 2018 Feb 1;140(2):0208011-9. doi: 10.1115/1.4037790. J Biomech Eng. 2018. PMID: 28857114 Free PMC article.
-
DNA stretching as a probe for nucleic acid interactions: Reply to Comments on "Biophysical characterization of DNA binding from single molecule force measurements" by Kathy R. Chaurasiya, Thayaparan Paramanathan, Micah J. McCauley, Mark C. Williams.Phys Life Rev. 2010 Sep 1;7(3):358-361. doi: 10.1016/j.plrev.2010.07.008. Phys Life Rev. 2010. PMID: 20922051 Free PMC article. No abstract available.
-
TAMRA-polypyrrole for A/T sequence visualization on DNA molecules.Nucleic Acids Res. 2018 Oct 12;46(18):e108. doi: 10.1093/nar/gky531. Nucleic Acids Res. 2018. PMID: 29931115 Free PMC article.
-
Revisiting blob theory for DNA diffusivity in slitlike confinement.Phys Rev Lett. 2013 Apr 19;110(16):168105. doi: 10.1103/PhysRevLett.110.168105. Epub 2013 Apr 19. Phys Rev Lett. 2013. PMID: 23679643 Free PMC article.
-
Nanochannel-Confined TAMRA-Polypyrrole Stained DNA Stretching by Varying the Ionic Strength from Micromolar to Millimolar Concentrations.Polymers (Basel). 2018 Dec 22;11(1):15. doi: 10.3390/polym11010015. Polymers (Basel). 2018. PMID: 30959999 Free PMC article.
References
-
- Rief M, Schaumann H, Gaub HE. Sequence dependent mechanics of single DNA molecules. Nat. Struct. Biol. 1999;6:346–349. - PubMed
-
- Cluzel P, Lebrum A, Heller C, Lavery R, Viovy J, Chatenay D, Caron F. DNA an extensible molecule. Science. 1996;271:792–794. - PubMed
-
- Smith S, Cui Y, Bustamante C. Overstretching B-DNA the elastic response of individual double stranded and single stranded DNA molecules. Science. 1996;271:795–799. - PubMed
-
- Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759–8770.

