Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1
- PMID: 20129251
- PMCID: PMC2818769
- DOI: 10.1016/j.ccr.2009.12.020
Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1
Abstract
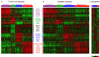
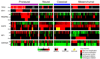
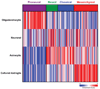
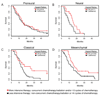
The Cancer Genome Atlas Network recently cataloged recurrent genomic abnormalities in glioblastoma multiforme (GBM). We describe a robust gene expression-based molecular classification of GBM into Proneural, Neural, Classical, and Mesenchymal subtypes and integrate multidimensional genomic data to establish patterns of somatic mutations and DNA copy number. Aberrations and gene expression of EGFR, NF1, and PDGFRA/IDH1 each define the Classical, Mesenchymal, and Proneural subtypes, respectively. Gene signatures of normal brain cell types show a strong relationship between subtypes and different neural lineages. Additionally, response to aggressive therapy differs by subtype, with the greatest benefit in the Classical subtype and no benefit in the Proneural subtype. We provide a framework that unifies transcriptomic and genomic dimensions for GBM molecular stratification with important implications for future studies.
Copyright (c) 2010 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
A simplified approach for molecular classification of glioblastomas (GBMs): experience from a tertiary care center in India.Brain Tumor Pathol. 2016 Jul;33(3):183-90. doi: 10.1007/s10014-016-0251-y. Epub 2016 Feb 10. Brain Tumor Pathol. 2016. PMID: 26865311
-
Molecular classification of glioblastoma based on immunohistochemical expression of EGFR, PDGFRA, NF1, IDH1, p53 and PTEN proteins.Pol J Pathol. 2021;72(1):1-10. doi: 10.5114/pjp.2021.106439. Pol J Pathol. 2021. PMID: 34060283
-
A glycolysis-based ten-gene signature correlates with the clinical outcome, molecular subtype and IDH1 mutation in glioblastoma.J Genet Genomics. 2017 Nov 20;44(11):519-530. doi: 10.1016/j.jgg.2017.05.007. Epub 2017 Sep 21. J Genet Genomics. 2017. PMID: 29169920
-
Isocitrate dehydrogenase status and molecular subclasses of glioma and glioblastoma.Neurosurg Focus. 2014 Dec;37(6):E13. doi: 10.3171/2014.9.FOCUS14505. Neurosurg Focus. 2014. PMID: 25434382 Review.
-
Glioblastoma: pathology, molecular mechanisms and markers.Acta Neuropathol. 2015 Jun;129(6):829-48. doi: 10.1007/s00401-015-1432-1. Epub 2015 May 6. Acta Neuropathol. 2015. PMID: 25943888 Review.
Cited by
-
Optimization of radiation dosing schedules for proneural glioblastoma.J Math Biol. 2016 Apr;72(5):1301-36. doi: 10.1007/s00285-015-0908-x. Epub 2015 Jun 21. J Math Biol. 2016. PMID: 26094055
-
Integrative functional genomics identifies RINT1 as a novel GBM oncogene.Neuro Oncol. 2012 Nov;14(11):1325-31. doi: 10.1093/neuonc/nos246. Epub 2012 Oct 16. Neuro Oncol. 2012. PMID: 23074196 Free PMC article.
-
Genetic and clinical characteristics of primary and secondary glioblastoma is associated with differential molecular subtype distribution.Oncotarget. 2015 Mar 30;6(9):7318-24. doi: 10.18632/oncotarget.3440. Oncotarget. 2015. PMID: 25821160 Free PMC article.
-
Identifying dysfunctional miRNA-mRNA regulatory modules by inverse activation, cofunction, and high interconnection of target genes: a case study of glioblastoma.Neuro Oncol. 2013 Jul;15(7):818-28. doi: 10.1093/neuonc/not018. Epub 2013 Mar 20. Neuro Oncol. 2013. PMID: 23516263 Free PMC article.
-
Sox11 expression in astrocytic gliomas: correlation with nestin/c-Met/IDH1-R132H expression phenotypes, p-Stat-3 and survival.Br J Cancer. 2013 May 28;108(10):2142-52. doi: 10.1038/bjc.2013.176. Epub 2013 Apr 25. Br J Cancer. 2013. PMID: 23619925 Free PMC article.
References
-
- Arjona D, Rey JA, Taylor SM. Early genetic changes involved in low-grade astrocytic tumor development. Curr Mol Med. 2006;6:645–650. - PubMed
-
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50CA58223/CA/NCI NIH HHS/United States
- CA127716/CA/NCI NIH HHS/United States
- U24CA126561/CA/NCI NIH HHS/United States
- U54HG003067/HG/NHGRI NIH HHS/United States
- P50 CA058223/CA/NCI NIH HHS/United States
- L30 CA104183-01/CA/NCI NIH HHS/United States
- R01 NS049720/NS/NINDS NIH HHS/United States
- U24 CA126561/CA/NCI NIH HHS/United States
- CA097257/CA/NCI NIH HHS/United States
- NS49720/NS/NINDS NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- P50 CA108961/CA/NCI NIH HHS/United States
- U54HG003273/HG/NHGRI NIH HHS/United States
- L30 CA104183-02/CA/NCI NIH HHS/United States
- U24CA126551/CA/NCI NIH HHS/United States
- U24CA126543/CA/NCI NIH HHS/United States
- U24CA126554/CA/NCI NIH HHS/United States
- U24CA126544/CA/NCI NIH HHS/United States
- U24 CA126551/CA/NCI NIH HHS/United States
- CA108961/CA/NCI NIH HHS/United States
- UL1 RR024992/RR/NCRR NIH HHS/United States
- U54 HG003273/HG/NHGRI NIH HHS/United States
- U54HG003079/HG/NHGRI NIH HHS/United States
- U24 CA126554/CA/NCI NIH HHS/United States
- P50 CA097257/CA/NCI NIH HHS/United States
- R01 CA127716/CA/NCI NIH HHS/United States
- U24 CA143848/CA/NCI NIH HHS/United States
- RR023248/RR/NCRR NIH HHS/United States
- U24 CA126543/CA/NCI NIH HHS/United States
- U24CA126563/CA/NCI NIH HHS/United States
- U24CA126546/CA/NCI NIH HHS/United States
- U54 HG003079/HG/NHGRI NIH HHS/United States
- K12 RR023248/RR/NCRR NIH HHS/United States
- U24 CA143867/CA/NCI NIH HHS/United States
- L30 CA104183/CA/NCI NIH HHS/United States
- F30 ES019449/ES/NIEHS NIH HHS/United States
- U24 CA126546/CA/NCI NIH HHS/United States
- U24 CA126563/CA/NCI NIH HHS/United States
- U24 CA126544/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

