The E6 protein from vaccinia virus is required for the formation of immature virions
- PMID: 20116821
- PMCID: PMC2830302
- DOI: 10.1016/j.virol.2010.01.012
The E6 protein from vaccinia virus is required for the formation of immature virions
Abstract
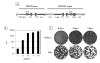
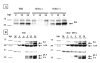
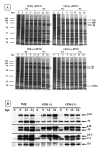
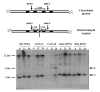
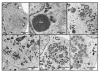
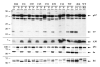
An IPTG-inducible mutant in the E6R gene of vaccinia virus was used to study the role of the E6 virion core protein in viral replication. In the absence of the inducer, the mutant exhibited a normal pattern DNA replication, concatemer resolution and late gene expression, but it showed an inhibition of virion structural protein processing it failed to produce infectious particles. Electron microscopic analysis showed that in the absence of IPTG viral morphogenesis was arrested before IV formation: crescents, aberrant or empty IV-like structures, and large aggregated virosomes were observed throughout the cytoplasm. The addition of IPTG to release a 12-h block showed that virus infectious particles could be formed in the absence of de novo DNA synthesis. Our observations show that in the absence of E6 the association of viroplasm with viral membrane crescents is impaired.
Published by Elsevier Inc.
Figures



Similar articles
-
Temperature-sensitive mutant in the vaccinia virus E6 protein produce virions that are transcriptionally inactive.Virology. 2010 Apr 10;399(2):221-30. doi: 10.1016/j.virol.2010.01.010. Epub 2010 Feb 8. Virology. 2010. PMID: 20116822 Free PMC article.
-
The vaccinia virus E6 protein influences virion protein localization during virus assembly.Virology. 2015 Aug;482:147-56. doi: 10.1016/j.virol.2015.02.056. Epub 2015 Apr 10. Virology. 2015. PMID: 25863879 Free PMC article.
-
Expression of the highly conserved vaccinia virus E6 protein is required for virion morphogenesis.Virology. 2009 Apr 10;386(2):478-85. doi: 10.1016/j.virol.2009.01.009. Epub 2009 Feb 12. Virology. 2009. PMID: 19217136 Free PMC article.
-
The major core protein P4a (A10L gene) of vaccinia virus is essential for correct assembly of viral DNA into the nucleoprotein complex to form immature viral particles.J Virol. 2001 Jul;75(13):5778-95. doi: 10.1128/JVI.75.13.5778-5795.2001. J Virol. 2001. PMID: 11390580 Free PMC article.
-
Vaccinia virus J1R protein: a viral membrane protein that is essential for virion morphogenesis.J Virol. 2002 Oct;76(19):9575-87. doi: 10.1128/jvi.76.19.9575-9587.2002. J Virol. 2002. PMID: 12208937 Free PMC article.
Cited by
-
From crescent to mature virion: vaccinia virus assembly and maturation.Viruses. 2014 Oct 7;6(10):3787-808. doi: 10.3390/v6103787. Viruses. 2014. PMID: 25296112 Free PMC article. Review.
-
Temperature-sensitive mutant in the vaccinia virus E6 protein produce virions that are transcriptionally inactive.Virology. 2010 Apr 10;399(2):221-30. doi: 10.1016/j.virol.2010.01.010. Epub 2010 Feb 8. Virology. 2010. PMID: 20116822 Free PMC article.
-
Vaccinia virus protein A3 is required for the production of normal immature virions and for the encapsidation of the nucleocapsid protein L4.Virology. 2015 Jul;481:1-12. doi: 10.1016/j.virol.2015.02.020. Epub 2015 Mar 9. Virology. 2015. PMID: 25765002 Free PMC article.
-
The vaccinia virus E6 protein influences virion protein localization during virus assembly.Virology. 2015 Aug;482:147-56. doi: 10.1016/j.virol.2015.02.056. Epub 2015 Apr 10. Virology. 2015. PMID: 25863879 Free PMC article.
-
Vaccinia virus mutations in the L4R gene encoding a virion structural protein produce abnormal mature particles lacking a nucleocapsid.J Virol. 2014 Dec;88(24):14017-29. doi: 10.1128/JVI.02126-14. Epub 2014 Sep 24. J Virol. 2014. PMID: 25253347 Free PMC article.
References
-
- Assarsson E, Greenbaum JA, Sundstrom M, Schaffer L, Hammond JA, Pasquetto V, Oseroff C, Hendrickson RC, Lefkowitz EJ, Tscharke DC, Sidney J, Grey HM, Head SR, Peters B, Sette A. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc.Natl.Acad.Sci.U.S.A. 2008;105:2140–2145. - PMC - PubMed
-
- Black EP, Moussatche N, Condit RC. Characterization of the interactions among vaccinia virus transcription factors G2R, A18R, and H5R. Virology. 1998;245:313–322. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

