Prevalent positive epistasis in Escherichia coli and Saccharomyces cerevisiae metabolic networks
- PMID: 20101242
- PMCID: PMC2837480
- DOI: 10.1038/ng.524
Prevalent positive epistasis in Escherichia coli and Saccharomyces cerevisiae metabolic networks
Abstract
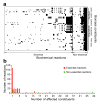
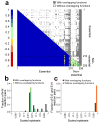
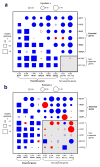
Epistasis refers to the interaction between genes. Although high-throughput epistasis data from model organisms are being generated and used to construct genetic networks, the extent to which genetic epistasis reflects biologically meaningful interactions remains unclear. We have addressed this question through in silico mapping of positive and negative epistatic interactions amongst biochemical reactions within the metabolic networks of Escherichia coli and Saccharomyces cerevisiae using flux balance analysis. We found that negative epistasis occurs mainly between nonessential reactions with overlapping functions, whereas positive epistasis usually involves essential reactions, is highly abundant and, unexpectedly, often occurs between reactions without overlapping functions. We offer mechanistic explanations of these findings and experimentally validate them for 61 S. cerevisiae gene pairs.
Conflict of interest statement
We have no competing financial interests.
Figures
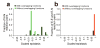
Comment in
-
Meaningful connections.Nat Rev Genet. 2010 Mar;11(3):172. doi: 10.1038/nrg2756. Nat Rev Genet. 2010. PMID: 21485431 No abstract available.
Similar articles
-
Epistatic interaction maps relative to multiple metabolic phenotypes.PLoS Genet. 2011 Feb 10;7(2):e1001294. doi: 10.1371/journal.pgen.1001294. PLoS Genet. 2011. PMID: 21347328 Free PMC article.
-
The causes of epistasis in genetic networks.Evolution. 2012 Feb;66(2):586-96. doi: 10.1111/j.1558-5646.2011.01451.x. Epub 2011 Sep 20. Evolution. 2012. PMID: 22276550
-
Epistatic interactions among metabolic genes depend upon environmental conditions.Mol Biosyst. 2014 Oct;10(10):2578-89. doi: 10.1039/c4mb00181h. Mol Biosyst. 2014. PMID: 25018101
-
Power graph compression reveals dominant relationships in genetic transcription networks.Mol Biosyst. 2013 Nov;9(11):2681-5. doi: 10.1039/c3mb70236g. Mol Biosyst. 2013. PMID: 23963601 Review.
-
A systems-biology approach to modular genetic complexity.Chaos. 2010 Jun;20(2):026102. doi: 10.1063/1.3455183. Chaos. 2010. PMID: 20590331 Free PMC article. Review.
Cited by
-
Yeast 5 - an expanded reconstruction of the Saccharomyces cerevisiae metabolic network.BMC Syst Biol. 2012 Jun 4;6:55. doi: 10.1186/1752-0509-6-55. BMC Syst Biol. 2012. PMID: 22663945 Free PMC article.
-
Transposon insertional mutagenesis of diverse yeast strains suggests coordinated gene essentiality polymorphisms.Nat Commun. 2022 Mar 21;13(1):1490. doi: 10.1038/s41467-022-29228-1. Nat Commun. 2022. PMID: 35314699 Free PMC article.
-
Smaller, scale-free gene networks increase quantitative trait heritability and result in faster population recovery.PLoS One. 2011 Feb 9;6(2):e14645. doi: 10.1371/journal.pone.0014645. PLoS One. 2011. PMID: 21347400 Free PMC article.
-
Evolution combined with genomic study elucidates genetic bases of isobutanol tolerance in Escherichia coli.Microb Cell Fact. 2011 Mar 25;10:18. doi: 10.1186/1475-2859-10-18. Microb Cell Fact. 2011. PMID: 21435272 Free PMC article.
-
Genome-wide association mapping reveals epistasis and genetic interaction networks in sugar beet.Theor Appl Genet. 2011 Jun;123(1):109-18. doi: 10.1007/s00122-011-1570-3. Epub 2011 Mar 30. Theor Appl Genet. 2011. PMID: 21448808
References
-
- Tischler J, Lehner B, Fraser AG. Evolutionary plasticity of genetic interaction networks. Nat Genet. 2008;40:390–1. - PubMed
-
- Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8:437–49. - PubMed
-
- Moore JH, Williams SM. Traversing the conceptual divide between biological and statistical epistasis: systems biology and a more modern synthesis. BioEssays. 2005;27:637–46. - PubMed
-
- Cordell HJ. Epistasis: what it means, what it doesn't mean, and statistical methods to detect it in humans. Hum Mol Genet. 2002;11:2463–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

