The genetic landscape of a cell
- PMID: 20093466
- PMCID: PMC5600254
- DOI: 10.1126/science.1180823
The genetic landscape of a cell
Abstract
A genome-scale genetic interaction map was constructed by examining 5.4 million gene-gene pairs for synthetic genetic interactions, generating quantitative genetic interaction profiles for approximately 75% of all genes in the budding yeast, Saccharomyces cerevisiae. A network based on genetic interaction profiles reveals a functional map of the cell in which genes of similar biological processes cluster together in coherent subsets, and highly correlated profiles delineate specific pathways to define gene function. The global network identifies functional cross-connections between all bioprocesses, mapping a cellular wiring diagram of pleiotropy. Genetic interaction degree correlated with a number of different gene attributes, which may be informative about genetic network hubs in other organisms. We also demonstrate that extensive and unbiased mapping of the genetic landscape provides a key for interpretation of chemical-genetic interactions and drug target identification.
Figures

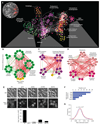
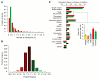
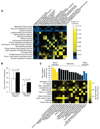

Comment in
-
Meaningful connections.Nat Rev Genet. 2010 Mar;11(3):172. doi: 10.1038/nrg2756. Nat Rev Genet. 2010. PMID: 21485431 No abstract available.
Similar articles
-
A global genetic interaction network maps a wiring diagram of cellular function.Science. 2016 Sep 23;353(6306):aaf1420. doi: 10.1126/science.aaf1420. Science. 2016. PMID: 27708008 Free PMC article.
-
Protein complexes are central in the yeast genetic landscape.PLoS Comput Biol. 2011 Feb;7(2):e1001092. doi: 10.1371/journal.pcbi.1001092. Epub 2011 Feb 24. PLoS Comput Biol. 2011. PMID: 21390331 Free PMC article.
-
An integrated approach to characterize genetic interaction networks in yeast metabolism.Nat Genet. 2011 May 29;43(7):656-62. doi: 10.1038/ng.846. Nat Genet. 2011. PMID: 21623372 Free PMC article.
-
Chemical-genetic approaches for exploring the mode of action of natural products.Prog Drug Res. 2008;66:237, 239-71. doi: 10.1007/978-3-7643-8595-8_5. Prog Drug Res. 2008. PMID: 18416308 Review.
-
Integrating genetic and protein-protein interaction networks maps a functional wiring diagram of a cell.Curr Opin Microbiol. 2018 Oct;45:170-179. doi: 10.1016/j.mib.2018.06.004. Epub 2018 Jul 28. Curr Opin Microbiol. 2018. PMID: 30059827 Free PMC article. Review.
Cited by
-
The proteome response to amyloid protein expression in vivo.PLoS One. 2012;7(11):e50123. doi: 10.1371/journal.pone.0050123. Epub 2012 Nov 21. PLoS One. 2012. PMID: 23185553 Free PMC article.
-
Quantitative genetic-interaction mapping in mammalian cells.Nat Methods. 2013 May;10(5):432-7. doi: 10.1038/nmeth.2398. Epub 2013 Feb 13. Nat Methods. 2013. PMID: 23407553 Free PMC article.
-
The roles of whole-genome and small-scale duplications in the functional specialization of Saccharomyces cerevisiae genes.PLoS Genet. 2013;9(1):e1003176. doi: 10.1371/journal.pgen.1003176. Epub 2013 Jan 3. PLoS Genet. 2013. PMID: 23300483 Free PMC article.
-
Analysis of genetic interaction networks shows that alternatively spliced genes are highly versatile.PLoS One. 2013;8(2):e55671. doi: 10.1371/journal.pone.0055671. Epub 2013 Feb 7. PLoS One. 2013. PMID: 23409018 Free PMC article.
-
Proteasome dysfunction triggers activation of SKN-1A/Nrf1 by the aspartic protease DDI-1.Elife. 2016 Aug 16;5:e17721. doi: 10.7554/eLife.17721. Elife. 2016. PMID: 27528192 Free PMC article.
References
-
- Dixon SJ, Costanzo M, Baryshnikova A, Andrews B, Boone C. Annu Rev Genet. 2009;43:601. - PubMed
-
- Waddington CH. The Strategy of the Gene. Allen & Unwin; London: 1957.
-
- Hartwell L. Science. 2004;303:774. - PubMed
-
- Tong AH, et al. Science. 2001;294:2364. - PubMed
-
- Tong AH, et al. Science. 2004;303:808. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

