The p40/ARPC1 subunit of Arp2/3 complex performs multiple essential roles in WASp-regulated actin nucleation
- PMID: 20071330
- PMCID: PMC2832997
- DOI: 10.1074/jbc.M109.054957
The p40/ARPC1 subunit of Arp2/3 complex performs multiple essential roles in WASp-regulated actin nucleation
Abstract
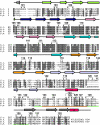
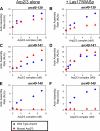
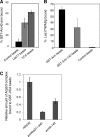
The Arp2/3 complex is a conserved seven-subunit actin-nucleating machine activated by WASp (Wiskott Aldrich syndrome protein). Despite its central importance in a broad range of cellular processes, many critical aspects of the mechanism of the Arp2/3 complex have yet to be resolved. In particular, some of the individual subunits in the complex have not been assigned clear functional roles, including p40/ARPC1. Here, we dissected the structure and function of Saccharomyces cerevisiae p40/ARPC1, which is encoded by the essential ARC40 gene, by analyzing 39 integrated alleles that target its conserved surfaces. We identified three distinct sites on p40/ARPC1 required for function in vivo: one site contacts p19/ARPC4, one contacts p15/ARPC5, and one site resides in an extended structural "arm" of p40/ARPC1. Using a novel strategy, we purified the corresponding lethal mutant Arp2/3 complexes from yeast and compared their actin nucleation activities. Lethal mutations at the contact with p19/ARPC4 specifically impaired WASp-induced nucleation. In contrast, lethal mutations at the contact with p15/ARPC5 led to unregulated ("leaky") nucleation in the absence of WASp. Lethal mutations in the extended arm drastically reduced nucleation, and the same mutations disrupted the ability of the purified p40/ARPC1 arm domain to bind the VCA domain of WASp. Together, these data indicate that p40/ARPC1 performs at least three distinct, essential functions in regulating Arp2/3 complex-mediated actin assembly: 1) suppression of spontaneous nucleation by the Arp2/3 complex, which requires proper contacts with p15/ARPC5; 2) propagation of WASp activation signals via contacts with p19/ARPC2; and 3) direct facilitation of actin nucleation through interactions of the extended arm with the VCA domain of WASp.
Figures


Similar articles
-
ARPC1/Arc40 mediates the interaction of the actin-related protein 2 and 3 complex with Wiskott-Aldrich syndrome protein family activators.J Biol Chem. 2004 Dec 24;279(52):54629-36. doi: 10.1074/jbc.M402357200. Epub 2004 Oct 14. J Biol Chem. 2004. PMID: 15485833
-
Identification of Wiskott-Aldrich syndrome protein (WASP) binding sites on the branched actin filament nucleator Arp2/3 complex.Proc Natl Acad Sci U S A. 2018 Feb 13;115(7):E1409-E1418. doi: 10.1073/pnas.1716622115. Epub 2018 Jan 31. Proc Natl Acad Sci U S A. 2018. PMID: 29386393 Free PMC article.
-
Interaction of cortactin and N-WASp with Arp2/3 complex.Curr Biol. 2002 Aug 6;12(15):1270-8. doi: 10.1016/s0960-9822(02)01035-7. Curr Biol. 2002. PMID: 12176354
-
Signalling to actin assembly via the WASP (Wiskott-Aldrich syndrome protein)-family proteins and the Arp2/3 complex.Biochem J. 2004 May 15;380(Pt 1):1-17. doi: 10.1042/BJ20040176. Biochem J. 2004. PMID: 15040784 Free PMC article. Review.
-
WASP family proteins, more than Arp2/3 activators.Biochem Soc Trans. 2016 Oct 15;44(5):1339-1345. doi: 10.1042/BST20160176. Biochem Soc Trans. 2016. PMID: 27911716 Free PMC article. Review.
Cited by
-
SOX10-regulated promoter use defines isoform-specific gene expression in Schwann cells.BMC Genomics. 2020 Aug 8;21(1):549. doi: 10.1186/s12864-020-06963-7. BMC Genomics. 2020. PMID: 32770939 Free PMC article.
-
Structure of Arp2/3 complex at a branched actin filament junction resolved by single-particle cryo-electron microscopy.Proc Natl Acad Sci U S A. 2022 May 31;119(22):e2202723119. doi: 10.1073/pnas.2202723119. Epub 2022 May 27. Proc Natl Acad Sci U S A. 2022. PMID: 35622886 Free PMC article.
-
Arp2/3 complex is bound and activated by two WASP proteins.Proc Natl Acad Sci U S A. 2011 Aug 16;108(33):E472-9. doi: 10.1073/pnas.1100236108. Epub 2011 Jun 15. Proc Natl Acad Sci U S A. 2011. PMID: 21676863 Free PMC article.
-
Dendritic spine formation and synaptic function require neurobeachin.Nat Commun. 2011 Nov 22;2:557. doi: 10.1038/ncomms1565. Nat Commun. 2011. PMID: 22109531 Free PMC article.
-
The Diverse Family of Arp2/3 Complexes.Trends Cell Biol. 2017 Feb;27(2):93-100. doi: 10.1016/j.tcb.2016.08.001. Epub 2016 Aug 29. Trends Cell Biol. 2017. PMID: 27595492 Free PMC article. Review.
References
-
- Stradal T. E., Rottner K., Disanza A., Confalonieri S., Innocenti M., Scita G. (2004) Trends Cell Biol. 14, 303–311 - PubMed
-
- Robinson R. C., Turbedsky K., Kaiser D. A., Marchand J. B., Higgs H. N., Choe S., Pollard T. D. (2001) Science 294, 1679–1684 - PubMed
-
- Goley E. D., Rodenbusch S. E., Martin A. C., Welch M. D. (2004) Mol. Cell 16, 269–279 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

