Alternative epigenetic chromatin states of polycomb target genes
- PMID: 20062800
- PMCID: PMC2799325
- DOI: 10.1371/journal.pgen.1000805
Alternative epigenetic chromatin states of polycomb target genes
Abstract
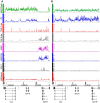
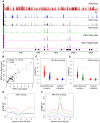
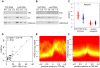
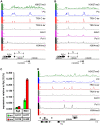
Polycomb (PcG) regulation has been thought to produce stable long-term gene silencing. Genomic analyses in Drosophila and mammals, however, have shown that it targets many genes, which can switch state during development. Genetic evidence indicates that critical for the active state of PcG target genes are the histone methyltransferases Trithorax (TRX) and ASH1. Here we analyze the repertoire of alternative states in which PcG target genes are found in different Drosophila cell lines and the role of PcG proteins TRX and ASH1 in controlling these states. Using extensive genome-wide chromatin immunoprecipitation analysis, RNAi knockdowns, and quantitative RT-PCR, we show that, in addition to the known repressed state, PcG targets can reside in a transcriptionally active state characterized by formation of an extended domain enriched in ASH1, the N-terminal, but not C-terminal moiety of TRX and H3K27ac. ASH1/TRX N-ter domains and transcription are not incompatible with repressive marks, sometimes resulting in a "balanced" state modulated by both repressors and activators. Often however, loss of PcG repression results instead in a "void" state, lacking transcription, H3K27ac, or binding of TRX or ASH1. We conclude that PcG repression is dynamic, not static, and that the propensity of a target gene to switch states depends on relative levels of PcG, TRX, and activators. N-ter TRX plays a remarkable role that antagonizes PcG repression and preempts H3K27 methylation by acetylation. This role is distinct from that usually attributed to TRX/MLL proteins at the promoter. These results have important implications for Polycomb gene regulation, the "bivalent" chromatin state of embryonic stem cells, and gene expression in development.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

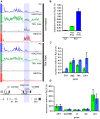
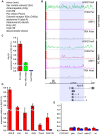
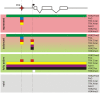
Similar articles
-
Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins.Genes Dev. 2006 Aug 1;20(15):2041-54. doi: 10.1101/gad.388706. Genes Dev. 2006. PMID: 16882982 Free PMC article.
-
Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II.PLoS Genet. 2008 Oct;4(10):e1000217. doi: 10.1371/journal.pgen.1000217. Epub 2008 Oct 10. PLoS Genet. 2008. PMID: 18846226 Free PMC article.
-
CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing.Development. 2009 Sep;136(18):3131-41. doi: 10.1242/dev.037127. Development. 2009. PMID: 19700617 Free PMC article.
-
Epigenetic inheritance of chromatin states mediated by Polycomb and trithorax group proteins in Drosophila.Prog Mol Subcell Biol. 2005;38:31-63. doi: 10.1007/3-540-27310-7_2. Prog Mol Subcell Biol. 2005. PMID: 15881890 Review.
-
Polycomb Recruiters Inside and Outside of the Repressed Domains.Int J Mol Sci. 2023 Jul 13;24(14):11394. doi: 10.3390/ijms241411394. Int J Mol Sci. 2023. PMID: 37511153 Free PMC article. Review.
Cited by
-
Inner workings and regulatory inputs that control Polycomb repressive complex 2.Chromosoma. 2012 Jun;121(3):221-34. doi: 10.1007/s00412-012-0361-1. Epub 2012 Feb 19. Chromosoma. 2012. PMID: 22349693 Free PMC article. Review.
-
An Ash1-Like Protein MoKMT2H Null Mutant Is Delayed for Conidium Germination and Pathogenesis in Magnaporthe oryzae.Biomed Res Int. 2016;2016:1575430. doi: 10.1155/2016/1575430. Epub 2016 Sep 26. Biomed Res Int. 2016. PMID: 27747223 Free PMC article.
-
Metazoan promoters: emerging characteristics and insights into transcriptional regulation.Nat Rev Genet. 2012 Mar 6;13(4):233-45. doi: 10.1038/nrg3163. Nat Rev Genet. 2012. PMID: 22392219 Review.
-
dBRWD3 Regulates Tissue Overgrowth and Ectopic Gene Expression Caused by Polycomb Group Mutations.PLoS Genet. 2016 Sep 2;12(9):e1006262. doi: 10.1371/journal.pgen.1006262. eCollection 2016 Sep. PLoS Genet. 2016. PMID: 27588417 Free PMC article.
-
Ash1 counteracts Polycomb repression independent of histone H3 lysine 36 methylation.EMBO Rep. 2019 Apr;20(4):e46762. doi: 10.15252/embr.201846762. Epub 2019 Mar 4. EMBO Rep. 2019. PMID: 30833342 Free PMC article.
References
-
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. - PubMed
-
- Poux S, Horard B, Sigrist CJA, Pirrotta V. The Drosophila Trithorax protein is a coactivator required to prevent re-establishment of Polycomb silencing. Development. 2002;129:2483–2493. - PubMed
-
- Smith ST, Petruk S, Sedkov Y, Cho E, Tillib S, et al. Modulation of heat shock gene expression by the TAC1 chromatin-modifying complex. Nat Cell Biol. 2004;6:162–167. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

