APOBEC3 proteins mediate the clearance of foreign DNA from human cells
- PMID: 20062055
- PMCID: PMC2921484
- DOI: 10.1038/nsmb.1744
APOBEC3 proteins mediate the clearance of foreign DNA from human cells
Abstract
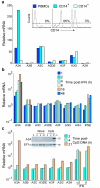
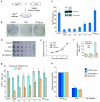
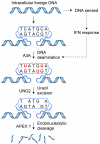
Bacteria evolved restriction endonucleases to prevent interspecies DNA transmission and bacteriophage infection. Here we show that human cells possess an analogous mechanism. APOBEC3A is induced by interferon following DNA detection, and it deaminates foreign double-stranded DNA cytidines to uridines. These atypical DNA nucleosides are converted by the uracil DNA glycosylase UNG2 to abasic lesions, which lead to foreign DNA degradation. This mechanism is evident in cell lines and primary monocytes, where up to 97% of cytidines in foreign DNA are deaminated. In contrast, cellular genomic DNA appears unaffected. Several other APOBEC3s also restrict foreign gene transfer. Related proteins exist in all vertebrates, indicating that foreign DNA restriction may be a conserved innate immune defense mechanism. The efficiency and fidelity of genetic engineering, gene therapy, and DNA vaccination are likely to be influenced by this anti-DNA defense system.
Figures


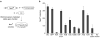
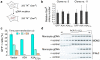
Comment in
-
Human cells clear foreign DNA.Nat Rev Genet. 2010 Mar;11(3):172. doi: 10.1038/nrg2757. Nat Rev Genet. 2010. PMID: 21485432 No abstract available.
Similar articles
-
Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A.J Biol Chem. 2012 Oct 5;287(41):34801-8. doi: 10.1074/jbc.M112.385161. Epub 2012 Aug 15. J Biol Chem. 2012. PMID: 22896697 Free PMC article.
-
Virion-associated uracil DNA glycosylase-2 and apurinic/apyrimidinic endonuclease are involved in the degradation of APOBEC3G-edited nascent HIV-1 DNA.J Biol Chem. 2007 Apr 20;282(16):11667-75. doi: 10.1074/jbc.M606864200. Epub 2007 Feb 1. J Biol Chem. 2007. PMID: 17272283
-
Strikingly different properties of uracil-DNA glycosylases UNG2 and SMUG1 may explain divergent roles in processing of genomic uracil.DNA Repair (Amst). 2012 Jun 1;11(6):587-93. doi: 10.1016/j.dnarep.2012.03.003. Epub 2012 Apr 6. DNA Repair (Amst). 2012. PMID: 22483865
-
Error-free versus mutagenic processing of genomic uracil--relevance to cancer.DNA Repair (Amst). 2014 Jul;19:38-47. doi: 10.1016/j.dnarep.2014.03.028. Epub 2014 Apr 18. DNA Repair (Amst). 2014. PMID: 24746924 Review.
-
[APOBEC3s: history of an antiviral and mutagenic protein family].Virologie (Montrouge). 2020 Dec 1;24(6):381-418. doi: 10.1684/vir.2020.0870. Virologie (Montrouge). 2020. PMID: 33441290 Review. French.
Cited by
-
Sustained high expression of multiple APOBEC3 cytidine deaminases in systemic lupus erythematosus.Sci Rep. 2021 Apr 12;11(1):7893. doi: 10.1038/s41598-021-87024-1. Sci Rep. 2021. PMID: 33846459 Free PMC article.
-
Evolution of the primate APOBEC3A cytidine deaminase gene and identification of related coding regions.PLoS One. 2012;7(1):e30036. doi: 10.1371/journal.pone.0030036. Epub 2012 Jan 17. PLoS One. 2012. PMID: 22272271 Free PMC article.
-
Mutation Processes in 293-Based Clones Overexpressing the DNA Cytosine Deaminase APOBEC3B.PLoS One. 2016 May 10;11(5):e0155391. doi: 10.1371/journal.pone.0155391. eCollection 2016. PLoS One. 2016. PMID: 27163364 Free PMC article.
-
Genome-wide alterations of uracil distribution patterns in human DNA upon chemotherapeutic treatments.Elife. 2020 Sep 21;9:e60498. doi: 10.7554/eLife.60498. Elife. 2020. PMID: 32956035 Free PMC article.
-
Molecular origins of APOBEC-associated mutations in cancer.DNA Repair (Amst). 2020 Oct;94:102905. doi: 10.1016/j.dnarep.2020.102905. Epub 2020 Jul 6. DNA Repair (Amst). 2020. PMID: 32818816 Free PMC article. Review.
References
Additional References for Online Methods
-
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. - PubMed
References
-
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. - PubMed
-
- Uematsu S, Akira S. Toll-like receptors and Type I interferons. J Biol Chem. 2007;282:15319–15323. - PubMed
-
- Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. - PubMed
-
- Roberts TL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. - PubMed
-
- Burckstummer T, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 AI064046/AI/NIAID NIH HHS/United States
- R01 AI064046-04/AI/NIAID NIH HHS/United States
- T32 CA009138/CA/NCI NIH HHS/United States
- R01 GM080437-04/GM/NIGMS NIH HHS/United States
- CA009138/CA/NCI NIH HHS/United States
- R56 AI064046/AI/NIAID NIH HHS/United States
- F32 GM090437/GM/NIGMS NIH HHS/United States
- R56 AI064046-06A1/AI/NIAID NIH HHS/United States
- R01 GM080437/GM/NIGMS NIH HHS/United States
- R01 AI064046-05S1/AI/NIAID NIH HHS/United States
- AI064046/AI/NIAID NIH HHS/United States
- R01 AI064046-05/AI/NIAID NIH HHS/United States
- R01 GM080437-04S1/GM/NIGMS NIH HHS/United States
- GM090437/GM/NIGMS NIH HHS/United States
- R01 AI064046/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

